Purdue Online Writing Lab Purdue OWL® College of Liberal Arts

Tables and Figures

Welcome to the Purdue OWL
This page is brought to you by the OWL at Purdue University. When printing this page, you must include the entire legal notice.
Copyright ©1995-2018 by The Writing Lab & The OWL at Purdue and Purdue University. All rights reserved. This material may not be published, reproduced, broadcast, rewritten, or redistributed without permission. Use of this site constitutes acceptance of our terms and conditions of fair use.
Note: This page reflects the latest version of the APA Publication Manual (i.e., APA 7), which released in October 2019. The equivalent resources for the older APA 6 style can be found at this page as well as at this page (our old resources covered the material on this page on two separate pages).
The purpose of tables and figures in documents is to enhance your readers' understanding of the information in the document; usually, large amounts of information can be communicated more efficiently in tables or figures. Tables are any graphic that uses a row and column structure to organize information, whereas figures include any illustration or image other than a table.
General guidelines
Visual material such as tables and figures can be used quickly and efficiently to present a large amount of information to an audience, but visuals must be used to assist communication, not to use up space, or disguise marginally significant results behind a screen of complicated statistics. Ask yourself this question first: Is the table or figure necessary? For example, it is better to present simple descriptive statistics in the text, not in a table.
Relation of Tables or Figures and Text
Because tables and figures supplement the text, refer in the text to all tables and figures used and explain what the reader should look for when using the table or figure. Focus only on the important point the reader should draw from them, and leave the details for the reader to examine on their own.
Documentation
If you are using figures, tables and/or data from other sources, be sure to gather all the information you will need to properly document your sources.
Integrity and Independence
Each table and figure must be intelligible without reference to the text, so be sure to include an explanation of every abbreviation (except the standard statistical symbols and abbreviations).
Organization, Consistency, and Coherence
Number all tables sequentially as you refer to them in the text (Table 1, Table 2, etc.), likewise for figures (Figure 1, Figure 2, etc.). Abbreviations, terminology, and probability level values must be consistent across tables and figures in the same article. Likewise, formats, titles, and headings must be consistent. Do not repeat the same data in different tables.
Data in a table that would require only two or fewer columns and rows should be presented in the text. More complex data is better presented in tabular format. In order for quantitative data to be presented clearly and efficiently, it must be arranged logically, e.g. data to be compared must be presented next to one another (before/after, young/old, male/female, etc.), and statistical information (means, standard deviations, N values) must be presented in separate parts of the table. If possible, use canonical forms (such as ANOVA, regression, or correlation) to communicate your data effectively.

A generic example of a table with multiple notes formatted in APA 7 style.
Elements of Tables
Number all tables with Arabic numerals sequentially. Do not use suffix letters (e.g. Table 3a, 3b, 3c); instead, combine the related tables. If the manuscript includes an appendix with tables, identify them with capital letters and Arabic numerals (e.g. Table A1, Table B2).
Like the title of the paper itself, each table must have a clear and concise title. Titles should be written in italicized title case below the table number, with a blank line between the number and the title. When appropriate, you may use the title to explain an abbreviation parenthetically.
Comparison of Median Income of Adopted Children (AC) v. Foster Children (FC)
Keep headings clear and brief. The heading should not be much wider than the widest entry in the column. Use of standard abbreviations can aid in achieving that goal. There are several types of headings:
- Stub headings describe the lefthand column, or stub column , which usually lists major independent variables.
- Column headings describe entries below them, applying to just one column.
- Column spanners are headings that describe entries below them, applying to two or more columns which each have their own column heading. Column spanners are often stacked on top of column headings and together are called decked heads .
- Table Spanners cover the entire width of the table, allowing for more divisions or combining tables with identical column headings. They are the only type of heading that may be plural.
All columns must have headings, written in sentence case and using singular language (Item rather than Items) unless referring to a group (Men, Women). Each column’s items should be parallel (i.e., every item in a column labeled “%” should be a percentage and does not require the % symbol, since it’s already indicated in the heading). Subsections within the stub column can be shown by indenting headings rather than creating new columns:
Chemical Bonds
Ionic
Covalent
Metallic
The body is the main part of the table, which includes all the reported information organized in cells (intersections of rows and columns). Entries should be center aligned unless left aligning them would make them easier to read (longer entries, usually). Word entries in the body should use sentence case. Leave cells blank if the element is not applicable or if data were not obtained; use a dash in cells and a general note if it is necessary to explain why cells are blank. In reporting the data, consistency is key: Numerals should be expressed to a consistent number of decimal places that is determined by the precision of measurement. Never change the unit of measurement or the number of decimal places in the same column.
There are three types of notes for tables: general, specific, and probability notes. All of them must be placed below the table in that order.
General notes explain, qualify or provide information about the table as a whole. Put explanations of abbreviations, symbols, etc. here.
Example: Note . The racial categories used by the US Census (African-American, Asian American, Latinos/-as, Native-American, and Pacific Islander) have been collapsed into the category “non-White.” E = excludes respondents who self-identified as “White” and at least one other “non-White” race.
Specific notes explain, qualify or provide information about a particular column, row, or individual entry. To indicate specific notes, use superscript lowercase letters (e.g. a , b , c ), and order the superscripts from left to right, top to bottom. Each table’s first footnote must be the superscript a .
a n = 823. b One participant in this group was diagnosed with schizophrenia during the survey.
Probability notes provide the reader with the results of the tests for statistical significance. Asterisks indicate the values for which the null hypothesis is rejected, with the probability ( p value) specified in the probability note. Such notes are required only when relevant to the data in the table. Consistently use the same number of asterisks for a given alpha level throughout your paper.
* p < .05. ** p < .01. *** p < .001
If you need to distinguish between two-tailed and one-tailed tests in the same table, use asterisks for two-tailed p values and an alternate symbol (such as daggers) for one-tailed p values.
* p < .05, two-tailed. ** p < .01, two-tailed. † p <.05, one-tailed. †† p < .01, one-tailed.
Borders
Tables should only include borders and lines that are needed for clarity (i.e., between elements of a decked head, above column spanners, separating total rows, etc.). Do not use vertical borders, and do not use borders around each cell. Spacing and strict alignment is typically enough to clarify relationships between elements.

Example of a table in the text of an APA 7 paper. Note the lack of vertical borders.
Tables from Other Sources
If using tables from an external source, copy the structure of the original exactly, and cite the source in accordance with APA style .
Table Checklist
(Taken from the Publication Manual of the American Psychological Association , 7th ed., Section 7.20)
- Is the table necessary?
- Does it belong in the print and electronic versions of the article, or can it go in an online supplemental file?
- Are all comparable tables presented consistently?
- Are all tables numbered with Arabic numerals in the order they are mentioned in the text? Is the table number bold and left-aligned?
- Are all tables referred to in the text?
- Is the title brief but explanatory? Is it presented in italicized title case and left-aligned?
- Does every column have a column heading? Are column headings centered?
- Are all abbreviations; special use of italics, parentheses, and dashes; and special symbols explained?
- Are the notes organized according to the convention of general, specific, probability?
- Are table borders correctly used (top and bottom of table, beneath column headings, above table spanners)?
- Does the table use correct line spacing (double for the table number, title, and notes; single, one and a half, or double for the body)?
- Are entries in the left column left-aligned beneath the centered stub heading? Are all other column headings and cell entries centered?
- Are confidence intervals reported for all major point estimates?
- Are all probability level values correctly identified, and are asterisks attached to the appropriate table entries? Is a probability level assigned the same number of asterisks in all the tables in the same document?
- If the table or its data are from another source, is the source properly cited? Is permission necessary to reproduce the table?
Figures include all graphical displays of information that are not tables. Common types include graphs, charts, drawings, maps, plots, and photos. Just like tables, figures should supplement the text and should be both understandable on their own and referenced fully in the text. This section details elements of formatting writers must use when including a figure in an APA document, gives an example of a figure formatted in APA style, and includes a checklist for formatting figures.
Preparing Figures
In preparing figures, communication and readability must be the ultimate criteria. Avoid the temptation to use the special effects available in most advanced software packages. While three-dimensional effects, shading, and layered text may look interesting to the author, overuse, inconsistent use, and misuse may distort the data, and distract or even annoy readers. Design properly done is inconspicuous, almost invisible, because it supports communication. Design improperly, or amateurishly, done draws the reader’s attention from the data, and makes him or her question the author’s credibility. Line drawings are usually a good option for readability and simplicity; for photographs, high contrast between background and focal point is important, as well as cropping out extraneous detail to help the reader focus on the important aspects of the photo.
Parts of a Figure
All figures that are part of the main text require a number using Arabic numerals (Figure 1, Figure 2, etc.). Numbers are assigned based on the order in which figures appear in the text and are bolded and left aligned.
Under the number, write the title of the figure in italicized title case. The title should be brief, clear, and explanatory, and both the title and number should be double spaced.
The image of the figure is the body, and it is positioned underneath the number and title. The image should be legible in both size and resolution; fonts should be sans serif, consistently sized, and between 8-14 pt. Title case should be used for axis labels and other headings; descriptions within figures should be in sentence case. Shading and color should be limited for clarity; use patterns along with color and check contrast between colors with free online checkers to ensure all users (people with color vision deficiencies or readers printing in grayscale, for instance) can access the content. Gridlines and 3-D effects should be avoided unless they are necessary for clarity or essential content information.
Legends, or keys, explain symbols, styles, patterns, shading, or colors in the image. Words in the legend should be in title case; legends should go within or underneath the image rather than to the side. Not all figures will require a legend.
Notes clarify the content of the figure; like tables, notes can be general, specific, or probability. General notes explain units of measurement, symbols, and abbreviations, or provide citation information. Specific notes identify specific elements using superscripts; probability notes explain statistical significance of certain values.

A generic example of a figure formatted in APA 7 style.
Figure Checklist
(Taken from the Publication Manual of the American Psychological Association , 7 th ed., Section 7.35)
- Is the figure necessary?
- Does the figure belong in the print and electronic versions of the article, or is it supplemental?
- Is the figure simple, clean, and free of extraneous detail?
- Is the figure title descriptive of the content of the figure? Is it written in italic title case and left aligned?
- Are all elements of the figure clearly labeled?
- Are the magnitude, scale, and direction of grid elements clearly labeled?
- Are parallel figures or equally important figures prepared according to the same scale?
- Are the figures numbered consecutively with Arabic numerals? Is the figure number bold and left aligned?
- Has the figure been formatted properly? Is the font sans serif in the image portion of the figure and between sizes 8 and 14?
- Are all abbreviations and special symbols explained?
- If the figure has a legend, does it appear within or below the image? Are the legend’s words written in title case?
- Are the figure notes in general, specific, and probability order? Are they double-spaced, left aligned, and in the same font as the paper?
- Are all figures mentioned in the text?
- Has written permission for print and electronic reuse been obtained? Is proper credit given in the figure caption?
- Have all substantive modifications to photographic images been disclosed?
- Are the figures being submitted in a file format acceptable to the publisher?
- Have the files been produced at a sufficiently high resolution to allow for accurate reproduction?
- PRO Courses Guides New Tech Help Pro Expert Videos About wikiHow Pro Upgrade Sign In
- EDIT Edit this Article
- EXPLORE Tech Help Pro About Us Random Article Quizzes Request a New Article Community Dashboard This Or That Game Popular Categories Arts and Entertainment Artwork Books Movies Computers and Electronics Computers Phone Skills Technology Hacks Health Men's Health Mental Health Women's Health Relationships Dating Love Relationship Issues Hobbies and Crafts Crafts Drawing Games Education & Communication Communication Skills Personal Development Studying Personal Care and Style Fashion Hair Care Personal Hygiene Youth Personal Care School Stuff Dating All Categories Arts and Entertainment Finance and Business Home and Garden Relationship Quizzes Cars & Other Vehicles Food and Entertaining Personal Care and Style Sports and Fitness Computers and Electronics Health Pets and Animals Travel Education & Communication Hobbies and Crafts Philosophy and Religion Work World Family Life Holidays and Traditions Relationships Youth
- Browse Articles
- Learn Something New
- Quizzes Hot
- This Or That Game New
- Train Your Brain
- Explore More
- Support wikiHow
- About wikiHow
- Log in / Sign up
- Education and Communications
- College University and Postgraduate
- Academic Writing
How to Cite a Graph in a Paper
Last Updated: March 18, 2024 Fact Checked
This article was co-authored by Megan Morgan, PhD . Megan Morgan is a Graduate Program Academic Advisor in the School of Public & International Affairs at the University of Georgia. She earned her PhD in English from the University of Georgia in 2015. There are 14 references cited in this article, which can be found at the bottom of the page. This article has been fact-checked, ensuring the accuracy of any cited facts and confirming the authority of its sources. This article has been viewed 296,382 times.
Sometimes you may find it useful to include a graph from another source when writing a research paper. This is acceptable if you give credit to the original source. To do so, you generally provide a citation under the graph. The form this citation takes depends upon the citation style used in your discipline. Modern Language Association (MLA) style is used by English scholars and many humanities disciplines, while authors working in psychology, the social sciences and hard sciences often use the standards of the American Psychological Association (APA). Other humanities specialists and social scientists, including historians, use the Chicago/Turabian style, and engineering-related fields utilize the standards of the Institute of Electrical and Electronics Engineers (IEEE). Consult your instructor before writing a paper to determine which citation style is required.
Citing a Graph in MLA Style

- For example, you might refer to a graph showing tomato consumption patterns this way: "Due to the increasing popularity of salsa and ketchup, tomato consumption in the US has risen sharply in recent years (see fig. 1)."

- Figures should be numbered in the order they appear; your first graph or other illustration is "Fig. 1," your second "Fig. 2," and so on.
- Do not italicize the word “Figure” or “Fig.” or the numeral.

- For example, “Fig. 1. Rise in tomato consumption in the US, 1970-2000...”

- “Fig. 1. Rise in tomato consumption in the US, 1970-2000. Graph from John Green...”

- You also italicize the title of a website, such as this: Graph from State Fact Sheets...

- “Fig. 1. Rise in tomato consumption in the US, 1970-2000. Graph from John Green, Growing Vegetables in Your Backyard', (Hot Springs: Lake Publishers, 2002).
- If the graph came from an online source, follow the MLA guidelines for citing an online source: give the website name, publisher, date of publication, media, date of access, and pagination (if any -- if not, type “n. pag.”).
- For example, if your graph came from the USDA website, your citation would look like this: “Fig. 1. Rise in tomato consumption in the US, 1970-2000. Graph from State Fact Sheets. USDA. 1 Jan 2015. Web. 4 Feb. 2015. n. pag.”

- Fig. 1. Rise in tomato consumption in the US, 1970-2000. Graph from John Green, Growing Vegetables in Your Garden , (Hot Springs: Lake Publishers, 2002), 43. Print." [6] X Research source
- If you give the complete citation information in the caption, you do not need to also include it in your Works Cited page.
Citing a Graph in APA Format

- For example, you could write: “As seen in Figure 1, tomato consumption has risen sharply in the past three decades.”

- Figures should be numbered in the order they appear; your first graph or other illustration is Figure 1 , the second is Figure 2 , etc.
- If the graph has an existing title, give it in “sentence case.” This means you only capitalize the first letter of the first word in the sentence, as well as the first letter after a colon.

- For example: Figure 1. Rise in tomato consumption,1970-2000.
- Use sentence case for the description too.

- If the graph you’re presenting is your original work, meaning you collected all the data and compiled it yourself, you don’t need this phrase.
- For example: Figure 1. Rise in tomato consumption,1970-2000. Reprinted from...

- For example: Figure 1. Rise in tomato consumption,1970-2000. Reprinted from Growing Vegetables in Your Backyard (p. 43),

- For example: Figure 1. Rise in tomato consumption,1970-2000. Reprinted from Growing Vegetables in Your Backyard (p. 43), by J. Green, 2002, Hot Springs: Lake Publishers.

- Figure 1. Rise in tomato consumption, 1970-2000. Reprinted from Growing Vegetables in Your Backyard (p. 43), by J. Green, 2002, Hot Springs: Lake Publishers. Copyright 2002 by the American Tomato Growers' Association. Reprinted with permission. [13] X Research source
Citing a Graph Using Chicago/Turabian Standards

- For example, “Fig. 1. Rise in tomato consumption..."

- Fig. 1. Rise in tomato consumption (Graph by American Tomato Growers' Association. In Growing Vegetables in Your Backyard . John Green. Hot Springs: Lake Publishers, 2002, 43). [18] X Research source
Citing a Graph in IEEE Format

- If this marks the first time you've used this source, assign it a new number.
- If you've already used this source, refer back to the original source number.
- In our example, let's say this is the fifth source used in your paper. Your citation, then, will begin with a bracket and then "5": "[5..."

- TOMATO CONSUMPTION FIGURES [5, p. 43].
- Be sure to list complete source information in your endnotes. [21] X Research source
Community Q&A
You Might Also Like

- ↑ https://owl.purdue.edu/owl/research_and_citation/mla_style/mla_formatting_and_style_guide/mla_tables_figures_and_examples.html
- ↑ https://research.moreheadstate.edu/c.php?g=610039&p=4234946
- ↑ https://otis.libguides.com/mla_citations/images
- ↑ https://owl.english.purdue.edu/owl/resource/747/14/
- ↑ https://aut.ac.nz.libguides.com/APA7th/figures
- ↑ https://www.lib.sfu.ca/help/cite-write/citation-style-guides/apa/tables-figures
- ↑ https://guides.himmelfarb.gwu.edu/c.php?g=27779&p=170358
- ↑ https://graduate.asu.edu/sites/default/files/chicago-quick-reference.pdf
- ↑ https://guides.unitec.ac.nz/chicagoreferencing/images
- ↑ https://owl.purdue.edu/owl/research_and_citation/chicago_manual_17th_edition/cmos_formatting_and_style_guide/general_format.html
- ↑ https://libguides.dickinson.edu/c.php?g=56073&p=360111
- ↑ https://guides.lib.monash.edu/c.php?g=219786&p=6610144
- ↑ https://owl.purdue.edu/owl/research_and_citation/ieee_style/tables_figures_and_equations.html
- ↑ https://www.york.ac.uk/integrity/ieee.html
About This Article

To cite a graph in MLA style, refer to the graph in the text as Figure 1 in parentheses, and place a caption under the graph that says "Figure 1." Then, include a short description, such as the title of the graph, and list the authors first and last name, as well as the publication name, with the location, publisher, and year in parentheses. Finish the citation with the page number and resource format, which might be print or digital. If you want to cite a graph in APA, Chicago, or IEEE format, scroll down for tips from our academic reviewer. Did this summary help you? Yes No
- Send fan mail to authors
Reader Success Stories
Lilian Sumole
Nov 4, 2020
Did this article help you?

Nov 5, 2016
Savannah Caceres
Mar 25, 2017
Tiffany Taylor
Mar 6, 2017
E. Almaslam
May 15, 2017

Featured Articles

Trending Articles

Watch Articles

- Terms of Use
- Privacy Policy
- Do Not Sell or Share My Info
- Not Selling Info
Don’t miss out! Sign up for
wikiHow’s newsletter
Advertisement

- Previous Article
- Next Article
1. INTRODUCTION
2. the citation graph: concepts and limits, 3. extending the citation graph, 4. data in the citation graph, 5. discussion, 6. related work, 7. conclusions and future work, acknowledgments, author contributions, competing interests, funding information, data availability, data citation and the citation graph.
- Cite Icon Cite
- Open the PDF for in another window
- Permissions
- Article contents
- Figures & tables
- Supplementary Data
- Peer Review
- Search Site
Peter Buneman , Dennis Dosso , Matteo Lissandrini , Gianmaria Silvello; Data citation and the citation graph. Quantitative Science Studies 2022; 2 (4): 1399–1422. doi: https://doi.org/10.1162/qss_a_00166
Download citation file:
- Ris (Zotero)
- Reference Manager
The citation graph is a computational artifact that is widely used to represent the domain of published literature. It represents connections between published works, such as citations and authorship. Among other things, the graph supports the computation of bibliometric measures such as h -indexes and impact factors. There is now an increasing demand that we should treat the publication of data in the same way that we treat conventional publications. In particular, we should cite data for the same reasons that we cite other publications. In this paper we discuss what is needed for the citation graph to represent data citation. We identify two challenges: to model the evolution of credit appropriately (through references) over time and to model data citation not only to a data set treated as a single object but also to parts of it. We describe an extension of the current citation graph model that addresses these challenges. It is built on two central concepts: citable units and reference subsumption. We discuss how this extension would enable data citation to be represented within the citation graph and how it allows for improvements in current practices for bibliometric computations, both for scientific publications and for data.
1.1. Citations and the Citation Graph
Citation is essential to the creation and propagation of knowledge and is a well-understood part of scholarship and scientific publishing. Citations allow us to identify the cited material, retrieve it, give credit to its creator, date it, and provide partial knowledge of its subject and quality.
Exploration of the graph to find publications of interest.
Tracking of authorship of papers: Citing and following citations is one way to attribute credit to authors and to keep up to date with the work of others.
Dissemination of research findings: The exploration of citations and cited authors enables the dispersed communities of researchers to share their findings and engage in discussions.
Computation of bibliometrics for the analysis of one researcher, venue, or publication impact in particular fields. The citation graph is the basis for nearly all the currently used bibliometrics, such as impact factor and h-index .
Throughout this paper, we refer to an idealized “citation graph” as though it were a real and unique digital artifact that represents papers and the citations between them. Of course, it is not unique: Various organizations have distinct implementations of it. Among these, we count: Google Scholar, the Microsoft Academic Graph (MAG) 1 , the Open Academic Graph (OAG) ( Tang et al., 2008 ), Semantic Scholar (SS) 2 , AMiner (AM) 3 , and PubMed 4 (this is more a linked collection of documents than a full-fledged citation graph), Scopus 5 , and the Web of Science 6 . These graphs differ in many aspects, such as their coverage, their being open- or closed-access, and their schema; but in all of these, the basic structure is a directed graph, in which the vertices represent publications and the edges represent citations from one publication to another ( Price, 1965 ).
Most of the information about papers is contained in annotations of the nodes. The edges are generally typed but not annotated (an exception is MAG, which carries context , as we discuss later). Although in early models, nodes only represented papers and the only edges were “cites” edges, recently, citation graphs have been extended with richer information ( Peroni & Shotton, 2020 ). These extensions may carry author nodes with a “wrote” edge to papers, journal/conference nodes with a “part of” edge from papers, and subject nodes with the corresponding edges. Although representations differ, the purpose is similar: to provide the services described above.
1.2. The Need for Data Citation
Scientific publications increasingly rely on curated databases, which are numerous, “populated and updated with a great deal of human effort” ( Buneman, Cheney et al., 2008 ), and at the core of current scientific research 7 . In this context, references to data are starting to be placed alongside traditional references. Hence, there has been a strong demand ( FORCE-11, 2014 ; CODATA-ICSTI Task Group on Data Citation Standards and Practices, 2013 ) to give databases the same scholarly status as traditional scientific works and to define a shared methodology to cite data. Scientific publishers (e.g., Elsevier, PLoS, Springer, Nature) have taken up data citation by instituting policies to include data citations in the reference lists.
The open research culture ( Nosek, Alter et al., 2015 ) is based on methods and tools to share, discover, and access experimental data. Moreover papers, journals, and articles should provide access to all the data that they use ( Cousijn, Feeney et al., 2019 ). Researchers and practitioners (e.g., journalists and data scientists) who make use of electronic data should be able to cite the relevant data as they would cite a document from which they had extracted information ( Cousijn, Kenall et al., 2017 ; Nature Physics Editorial, 2016 ). As we shall see, the citation graphs can become a fundamental tool in the pursuit of the goal of accessibility and networking between papers and data.
We also observe that data occupy a crucial role today in research, emerging as a driving instrument in science ( Candela, Castelli et al., 2015 ). Data citations should be given the same scholarly status as traditional citations and contribute to bibliometrics indicators ( Belter, 2014 ; Peters, Kraker et al., 2016 ). Principles such as Findability, Accessibility, Interoperability, and Reusability (FAIR) ( Wilkinson, Dumontier et al., 2016 ) require data to be easily findable and accessible, qualities that are more readily available once data can be appropriately cited. In this sense, we can say that the FAIR principles encourage the adoption of data citation.
The reasons given for data citation are the same those given for a conventional citation ( CODATA-ICSTI Task Group on Data Citation Standards and Practices, 2013 ): recognition of the source (e.g., a title); credit for the author, curator, or agent; establishment of its currency (when it was created); where it was located; and how it was extracted. The last three of these fall under the general heading of provenance and are important when one wants to reproduce some analysis on the data or establish the trustworthiness of a claim.
Data sets and databases are usually more complex and varied than textual documents, and they introduce significant challenges for citation ( Silvello, 2018 ). Text publications have a fixed form, do not change over time, are interpretable as independent units, share a standard format and representation model, and are composed of predetermined, albeit domain-dependent, sets of elements that are considered as citable (e.g., the whole paper or book or a chapter). Scientific databases are structured according to diverse data models and accessed with a variety of query languages. What can be cited may range from a single datum to data subsets or aggregations specified by the person or agent that extracts the relevant data, and deciding a priori what can and cannot be cited is rarely feasible. Data citation introduces multiple citation types, besides the classical papers citing papers. These are papers citing data, data citing papers, and data citing data.
1.3. Data Citation in the Citation Graph
Our purpose in this paper is to discuss whether, in its current form, the current model of the citation graph can properly accommodate data citation. We claim that, despite all the features and modifications that have been added to various implementations of the citation graph, at least two significant features are generally missing or poorly represented. These shortcomings already limit what we can represent with existing implementations, and we argue that they make impossible the proper representation of data citations.
The first shortcoming concerns the assignment of credit when a referenced scientific work is corrected or augmented with another version. A typical example case is that of a preprint paper that gets cited before its peer-reviewed version is published. It is common for the authors to prefer that the preprint citations are merged with those of the peer-reviewed version. Something similar happens also when an updated version of a data set is published.
In the case of data, we need to consider that a database may be composed of multiple independently citable parts (e.g., a single record, a table, a view). Every single citable part can evolve and change over time and obtain citations (also views or downloads, when monitoring other scientometrics signals) at a different point in time. Therefore, it can be necessary to aggregate these statistics over all the versions of the same part to measure its impact and that of the database. The MAG and S2ORC databases have also an explicit notion of multiple versions of a paper, for example preprints and final published versions. It is however uncommon to “move” citations from one version to another, following some criteria or algorithm to correctly allocate citations. Yet, aggregating citation to a single version of a scientific work would have, among other things, the desirable effect of allowing proper evaluation of the impact of the work.
The second feature is the representation of context of a citation. Context is required for various reasons. It is typically used to describe the relevant part (e.g., page number) of a cited document. It may also carry, as in MAG, the surrounding text within the citing document helping to understand the reason for the citation; for example, a simple mention, a confutation, or a validation, such as those described in the OpenCitations ontology ( Daquino, Peroni et al., 2020 ). In the case of data citations, the context can contain the query identifying the cited data, expressed in different format (e.g., a URL, a filename, a SQL or SPARQL query, etc.). Despite a great deal of attention dedicated to the citation context—see, for instance, the Citation Context Analysis (CCA) discussed as early as the 1980s ( Freeman, Ding, & Milojevic, 2013 )—there is no systematic approach to representing it within citation graphs.
In fact, none of the largest citation-based systems, such as Scopus, MAG, and Google Scholar, properly take into account scientific databases as objects for use in the research literature. Google Data Search 8 allows us to search for indexed data sets, but it does not keep track of the citations to data or other types of statistics, such as clicks or downloads. Web of Science is one notable exception because it models data citations, even though only at the database level, via the Data Citation Index (DCI), now maintained by Clarivate Analytics ( Force, Robinson et al., 2016 ). Note that DCI is not publicly available and the data sets are indexed after a validation process.
Another effort is the Scholix framework ( Burton, Koers et al., 2017 ), which can be regarded as a set of guidelines and lightweight models that can be quickly adopted and expanded to facilitate interoperability among link providers. Finally, an example of an initiative that includes data and databases among the entities of the graph is the OpenAIRE Research Graph Data Model ( Manghi, Bardi et al., 2019 ), which leverages the OpenAIRE services to populate a research graph whose nodes include scientific results, organizations, funding agencies, communities, and data sources.
The conventional approach is to treat a data set as a single entity, in the same way, one would treat a scientific publication. However, this is far from ideal as typically only a small part of the data set or database is cited, and the authorship—the people who have contributed to the database—can vary widely with the part of the database being cited ( Buneman, Davidson, & Frew, 2016 ).
In this paper, we discuss the extension of the current model to enable the proper inclusion of data citations in the citation graph; and we discuss the evolution of a database: What happens to citations when new versions of the database appear? For the versioning issue, we describe a relation between scientific works (either papers or data) called subsumption . Through different policies, this relationship models effectively how credit should be transferred through time when updated versions of data appear in the graph. Finally, we discuss how to introduce data in the citation graph, considering the most common data citation strategies currently used in the world of research. In particular, we take inspiration from one of the solutions proposed by the Research Data Alliance ( RDA ) 9 . The RDA is a community-driven initiative launched in 2013 by different commissions. One of its working groups, the “Working Group on Data Citation: Making Dynamic Data Citable” (WGDC), has as one of its goals the identification and citation of arbitrary views of data. As a potential solution, the WGDC recommends an identification method based on PIDs assigned to queries.
The focus of this work is on data citation; but to ease the comprehension of the paper, we first discuss the limitations of the citation graph and the possible extensions we propose by focusing on textual documents, and then we extend the reasoning to data citation.
The paper is organized as follows: Section 2 describes some preliminary concepts and the limits of the citation graphs; in Section 3 we discuss the proposed solutions for the first three issues; Section 4 presents the proposed solution for the introduction of data in the citation graph; Section 5 sums up our main proposals and discusses possible lines of research and development; Section 6 describes the related work; finally, Section 7 presents conclusions and future work.
2.1. Core Concepts
2.1.1. citable unit.
By citable unit (CU), we mean a published entity—be it a paper, a chapter, or portion of data—which presents all the qualities necessary to be considered as a “citable work.” The characterization of a CU that we use, given in Wilke (2015) , requires that: it must be uniquely and unambiguously identifiable and citable; it must be available in perpetuity and in unchanged form; it must be accessible ; and it must be self-contained and complete . Self-contained and complete means that whatever new contribution is contained inside the piece of work, that contribution needs to be fully and clearly explained. This is not always the case for certain publications. Consider the slides of a scientific presentation. As they are used merely as a support for the oral presentation, they often cannot be fully understood without the corresponding talk. Also, the combination slides/registration of the talk may be incomplete, as many presenters tend to skip technical details during their presentations, referring to the complete published work.
Although some of these requirements are subjective, and not straightforward in databases, they still provide a workable starting point. The requirement that is most problematic for databases is that the citable unit must be unchanged . Databases evolve rapidly, and creating a citable unit for each version may be counterproductive. This is something we address in Section 4.2. Generally, what constitutes a citable unit is decided by convention. We should also note that some citable units comprise other citable units. The proceedings of a conference may be cited as may be a book on a topic whose chapters are written by different people and may also be individually cited. There is thus a “part-of” relationship between CUs that we discuss later.
In ( Daquino et al., 2020 ) a similar concept, bibliographic resource , is defined as a resource that cites and can be cited by other resources.
2.1.2. Reference
At the end of this paper, there is a list of references. Traditionally, a reference is a pointer to, and a brief description of, another publication in the literature. It is a short text composed of fields such as title, authors, year, venue, and others, that enables us to identify and find the entity (i.e., a paper, a book, or a survey) being referenced. Depending on aspects of the citing CU’s nature, like its field of research, the publication venue, or even language, different attributes of the reference may vary such as the format or the fields composing the reference. In physics, for example, titles are often omitted.
The important point is that, apart from the stylistic rendition of the reference, its contents are determined by the cited CU; hence, to within stylistic variations, the reference to a CU will be the same in any paper. In this paper, the reference determines the existence of a directed edge between two CUs: the citing and the cited one.
2.1.3. Citation
There is no universal agreement on the distinction between reference and citation , and the two terms are often used interchangeably ( Altman & Crosas, 2014 ; Daquino et al., 2020 ; Osareh, 1996 ; Price & Richardson, 2008 ).
One distinction proposed in Gilbert and Woolgar (1974) is that “reference” refers to the works mentioned in the reference section or bibliography of a paper. A reference may be mentioned once or many times in an article. Each of these mentions is considered a citation.
The distinction is crucial to our understanding of the citation graph. If we look at what goes in the body of a paper, we may find, for example, “Austen, J. (2004). pp 101–104.” We note that this textual artifact contains two parts. The first one is “Austen, J. (2004),” which we call a reference pointer . A reference pointer is, in general, a textual means that is used to denote a single bibliographic reference in the reference section when mentioned in the body of a paper. The second part of the citation is composed of some additional information, in this case “pp 101–104,” which may help the reader locate specific information within the cited paper. Note that the same reference pointer can occur several times in a paper and may have differing additional information, such as “pp 10–25” and “pp 110–120.”
Therefore, we can say that a citation is composed of the combination of the reference pointer with the (optional) information added to it in the paper’s body. The optional information in the paper’s body may be referred to as a form of context for the citation. This implies that there is a many-one relationship between citations and references, a fact that is supported by some discussions on the topic, for example “… the second necessary part of the citation or reference is the list of full references, which provides complete, formatted detail about the source, so that anyone reading the article can find it and verify it.” ( Wikipedia, 2021 ).
2.1.4. Reference annotation
We shall call this extra information, such as “pp 101–104,” reference annotation . In this paper, the reference annotation consists of all the information added to a reference pointer to qualify how it is used. This information is not part of the reference and can change depending on how that particular resource is used.
The Citation Typing Ontology ( Shotton, 2010 ) is replete with examples of other kinds of annotations such as “refutes,” or “ridicules,” which are clearly about the relationship between the citing and cited documents. In the Microsoft Academic Graph ( Sinha, Shen et al., 2015 ), the context —the text surrounding a citation in the source document—may be recorded as another form of annotation. The OpenCitations ontology ( Daquino et al., 2020 ) contains a class called annotation 10 attached to the in-text citation and to a reference which has a similar role. Here, we do not need to distinguish between the context of a reference pointer and its reference annotation: For our purposes these two concepts are the same, however it may be that certain applications will require some finer distinctions.
These definitions differ slightly from those in Daquino, Peroni, and Shotton (2018) and Daquino et al. (2020) , where a reference (called a bibliographic reference) and a reference pointer are manifestations of a citation. Moreover, in our example, the part “pp. 101–104” is a reference annotation, whereas in Daquino et al. (2020) it is a specialization of the citation. We do not specifically model the concept of specialization, as it can be inferred from the content of the reference annotation. Also, in Daquino et al. (2020) the pointer may include additional information, but the citation does not.
Summing up, we consider a reference annotation as a “box” that can contain information derived from the context of a reference pointer.
Generally speaking, the Citation Context Analysis (CCA), whose basis was first developed in the early 1980s, is the syntactic and semantic analysis of citation content, used to analyze the context of research behavior ( Freeman et al., 2013 ). CCA has been used as a promising addition to traditional quantitative citation analysis methods. One of the main aspects of CCA is that it incorporates qualitative factors, such as how one cites. In Daquino et al. (2020) this idea is captured by the concept of citation function , which is the function or purpose of the citation (e.g., to cite as background, extend, agree with the cited entity) to which each in-text reference pointer relates. In our proposal, this qualitative factor, or citation function, can be located in the reference annotation, and it could be inferred from the context of the reference pointer.
Even in a citation graph that represents conventional citations it is necessary to be able to attach information to a reference to create proper citations. Yet, in some citation graph implementations, this is impossible, because the reference relationship is represented as a directed but unannotated edge. As noted above, an exception is the Microsoft Academic Graph, which contains two kinds of edges between publications: unannotated edges and edges annotated with context. The reason for this omission may be the difficulty of collecting the relevant information; it may also be that it is not needed in the computation of most bibliometrics.
2.1.5. Part-of
The part-of relationship exists between two citable units in the graph; it describes the situation where one citation unit is somehow “contained” in the other. This is the case of papers published in an instance of a venue (e.g., the 2020 version of the ACM SIGMOD), and these issues being part of the venues themselves (e.g., ACM SIGMOD). This information is present for example in databases such as MAG and AMiner.
In the case of data, the part-of relationship is particularly important. Many databases and data sets have a hierarchical structure and may be cited at different levels of detail.
2.1.6. Database categories and citation
Static databases , which are used to support claims in a publication. These are typically “one-off” results of a set of experiments. For these databases, systems such as Mendeley 11 store data alongside the publication, so that a citation to the publication also serves as a citation to the data. Data journals ( Candela et al., 2015 ) (i.e., journals publishing papers describing data sets) are also employed as proxies to cite static data sets.
Evolving databases of source data such as weather data ( Philipp, Bartholy et al., 2010 ) or satellite image data ( Shanableh, Al-Ruzouq et al., 2019 ) that are collected for a wide range of purposes. Zenodo 12 , like Mendeley, stores data together with its representative publication. However, a publication about a data set and the data set itself can also have separate and unrelated DOIs. In this case the citation to the publication and to the database are distinguished. Moreover, it allows multiple versions of the same database to be deposited, with new DOIs for each one, thus keeping track of usage statistics like the number of downloads and views on each version. A citation to the database, or even to a document that describes the whole database, is generally regarded as inadequate. Usually, only a portion is used; hence, one needs to know the part (the sensor, the location of the image, or the time range) from which the data was extracted.
Finally, we have curated databases . These have largely replaced conventional biological reference works ( Buneman et al., 2008 ), and like the works they replace, involve substantial human effort. One advantage is that they are readily accessible and easy to search. Moreover, there are few limits on their size and complexity, and they can evolve rapidly with the subject matter. For these, the citation is a complex issue but it is just as crucial for curated databases as it is for the reference works that they replace.
The distinction between these three categories is not sharp, and there are many examples that lie in the overlap. For example, most source data databases involve a degree of curation.
2.2. Existing Limitations of the Citation Graph
Although implementations of the citation graph differ, the basic model consists of a directed graph 𝒢 = ( V , E ), where V is the set of papers and E ⊆ V × V is the set of directed edges corresponding to the citations among them: An edge 〈 p 1 , p 2 〉 connects the papers p 1 and p 2 , if p 1 cites p 2 . The following limitations of this simple model are obstacles to the representation of data citation, but can already be seen in conventional citations to papers.
2.2.1. Lack of context
Although in the basic model of the citation graph the nodes often contain information such as the title , the list of authors , or the venue of publication, it is lacking the information about the context of the citation, that is, all that kind of information that could be inferred from the context of the reference pointers, such as the specialization of the citation or the citation function. The only information provided by the edge 〈 p 1 , p 2 〉 is that p 1 cites p 2 , but it does not specify the why or the how of this citation. In the literature, we find the contextual citation graphs , which make apparent the textual contexts of each citation ( Bird, Dale et al., 2008 ; Daquino et al., 2020 ; Lo, Wang et al., 2019 ). These graphs contain information about reference annotations, which is what, in this work, we consider as the citation context.
Note that a lack of citation context is an issue that is related to not only data citation but the whole scientific citation infrastructure and ecosystem. How one document is cited in another, whether cited as a piece of evidence or a tool, could greatly influence how the scientific bibliographic universe is built and how credit should be assigned between researchers.
2.2.2. Versions
Ideally, the papers in the citation graph should only cite papers in the past (i.e., papers that already exist when the new paper is introduced in the graph [ Lo et al., 2019 ]). If this is the case, the citation graph is a DAG (Directed Acyclic Graph).
However, this often is not true because some of the papers in V go through revisions and modifications. This happens for many reasons and with many variations. Among the possible cases: It may be that several copies of one work are to be found on the internet; that one version is an “abstract” and is published in some conference proceedings, and a “full version” is later published in some journal; or that one version is published in some archive online and then a fully fledged paper is released in a conference or journal.
To receive credit, it is generally in the authors’ interest to have these documents seen as one. What appears to happen in Google Scholar, for example, is that all versions are clustered together, and one of them, the “main” version, is selected to be the recipient of all references.
Consider the following situation: document A is published, and a document P citing A is subsequently published. Document B, a revision and possibly an extension of A, is then published, taking A’s place in the graph. If this new version B contains new outgoing citations to P, then a cycle is created, and the graph is no longer a DAG (P → A ⇝ B → P). This problem may be solved by separating A and B.
Another source for cycles in citation graphs that cannot be avoided are papers by the same authors created at the same time (e.g., a full paper written together with a demo paper or extended abstract). In this case, the problem can be solved, for example, by conflating the papers.
Another problem arises when the system, for some reason, decides that B becomes the “main” representative of the publication. In this case, what happens with services such as Google Scholar is that the references first given to A are rerouted to B. This can be confusing as the reference annotation (e.g., the page number) may no longer be valid.
2.2.3. Citations to data
One of the primary roles of data citation is to give credit and attribution to the work of data creators and curators ( CODATA-ICSTI Task Group on Data Citation Standards and Practices, 2013 ). If integrated into the citation graph data citations can be represented and analyzed as if they were conventional citations, with data CUs and corresponding authors receiving citations and thus credit for their work. However, services such as Google Scholar or Scopus do not allow databases into their citation graph.
Data journals ( Candela et al., 2015 ) enable the publication of papers describing a database that works as a proxy for it and its authors and receives its citations. This is a possible solution, but it is not complete as it does not consider citations referring to general queries.
To give appropriate credit to the contributors to the various parts of a complex curated database, one approach to data citation ( Buneman, Christie et al., 2020 ) is to automatically create short papers, citation summaries , for each citable part of the database and publish them in a dedicated online journal. This enables the contributors to receive proper bibliometric credit for their contributions to the database. In this approach, a new summary for a view is generated whenever that view changes substantially. This summary can then be included in the current implementations of citation graphs and receive citations.
To conclude, unless there is some form of representation of the cited database or the cited query in the form of a paper or journal, current citation graphs do not include databases as nodes and citations to data as edges.
We describe two key extensions to the citation graph needed to deal with both the structural complexity and evolution of databases. These extensions already exist in a limited form in some implementations of the citation graph. However, we need to specify them precisely and understand how they help with the limitations described above and with data citations. What we propose is independent of any specific implementation of the citation graph and, for the most part, it can be incorporated as extensions to those implementations rather than requiring a completely new implementation of the supporting database.
3.1. Reference Annotation
As discussed above, a reference is represented by an edge in the citation graph. However, to represent a citation accurately, we need to add reference annotations . That is, we need to annotate the edges. Unfortunately, most data models currently implemented do not support data on edges 13 , so for consistency with these models, our diagrams include a new kind of node rather than a new kind of edge.
Consider Figure 1 . Two papers, P 1 and P 2 , are represented with circular nodes. We use these nodes to represent citable units. They are annotated with all the information that usually constitutes one reference, such as title, authors, year of publication, journal name, and DOI.

Use of references and reference annotations. Each reference is an edge connecting one citing unit to the cited one, and, if it exists, it is unique. One reference may have one or more reference annotations, each giving rise to a citation.
In this example P 1 references P 2 . We can imagine the reference appearing in the “References” section of P 1 as something similar to “Johansson, L. C. et al. (2019). XFEL structures of the human MT 2 melatonin receptor reveal the basis of subtype selectivity. Nature, 569(7755), 289–292. doi: 10.1038/s41586-019-1144-0.” The use of this reference in the paper is reflected by the presence of the reference edge between P 1 and P 2 and the reference node reference_1 . This is a different kind of node, which contains information such as the edge type (reference), the timestamp of when the citation was registered by the system and the type of reference (in this case from a paper to another paper). The actual information contained by the node can be modeled according to whatever model we decide to follow (e.g., the aforementioned Open Citation ontology).
Suppose now that P 1 cites P 2 twice. Each time, it does not merely refer to the whole paper P 2 , but specific parts of it. The node reference_1 has two other neighbor nodes, called reference annotation nodes , ra_1 and ra_2 . These two nodes contain the information describing the reference annotations found in P 1 used to cite P 2 , such as the context, references to particular tables or images, comment on the nature of the citation (e.g., that the authors of P 1 agree or disagree with P 2 ). In the example, these annotations carry page numbers. Hence, the combination of reference_1 with ra_1 makes one citation.
Reference and reference annotation nodes are the addition that we make to the citation graph to face the first problem.
3.2. Subsumption
Often new documents take the place of older versions, becoming also the recipients of both new and old citations. This behavior is handled behind the scenes by some existing implementations of the citation graph (notably Google Scholar). To deal with this phenomenon transparently, we propose the introduction in the citation graph of a new relation, called subsumes .
In Figure 2 we see a situation similar to the one of Figure 1 , where P 1 is citing P 2 at time 1. Now, imagine that a new version of the same paper, P 2 ′ , is published and inserted in the citation graph at time 2. The reference for P 2 ′ should also have a version number or something that distinguishes it from P 2 . The relation subsumes between P 2 ′ and P 2 indicates that the former is a new version of the latter, and is, from now on, the paper to consult and reference.

The “subsumes” relation between two CUs.
In some scientific areas, a journal “paper” P 2 ′ may be treated as a version of an earlier conference “abstract” P 2 , even though the two differ substantially. Because of this we do not want to destroy the original link from P 1 to P 2 ; to do so would be to “rewrite history” and remove information from the graph, and we strongly feel this should not be the case with the citation graph. The subsumes relation is present to indicate that one paper is a version of another and, crucially, that author credit can be transferred from the subsuming paper to the subsumed paper. On the one hand, the transfer of credit enables a more comprehensive measure of author contributions (e.g., increasing the number of citations on the latest version of the publication). On the other hand, credit transfer also transparently reflects the impact that the publication, seen as the aggregation of its different versions, has on the research community. Different types of subsumption can be defined, such as the kind of subsumption that propagates the citations to the single papers to their journal, thus computing its impact factor.
It would be wrong to transfer the credit for writing a paper to more than one other paper, so the subsumption relation is many-one. It is necessarily acyclic, thus it is a forest with the roots of the trees in that forest being the papers that are designated to receive the credit. It may be useful to have a term for a root node on the subsumption graph, perhaps primary citable units (PCU). It is interesting to note a similar approach in the MAG 14 , which lists the CU P 2 under the PCU P 2 ′ , keeps the citation count for P 2 and P 2 ′ distinct, and reports, for example, “124 citations” for P 2 , “325 citations” for P 2 ′ but adds, to P 2 ′ , “449 citations for all.”
Here we discuss how we place databases in the citation graph. We shall find that the two extensions we have discussed—edge annotation and subsumption—are essential to accommodating databases. In particular, they allow us to deal with databases, which tend to be updated and thus change much more frequently than papers. We could treat each version or instance of the database as a distinct document, but—at least for author credit—this would be a limitation, if not counterproductive.
First of all, we use the term “database” in the most general sense to refer to a conventional relational database, an ontology, some form of graph database, or a database that is a collection of files ( Buneman et al., 2016 ). One might then say that anything one has termed a database is a citable unit. The problem is that parts of the database may also be citable units. The reason we need to discuss parts of the database is twofold: First, where in the database one finds something is, like page numbers, a form of location or partial provenance; the second authorship may vary with what part of the database is being cited ( Buneman et al., 2016 ).
With “part” of a database we intend a view ( Buneman et al., 2016 ). A view is a query which we again generalize to being anything from a relational query for a relational database, a directory path or URI for a collection of files, or some query in one of the several languages that have been developed for ontologies and graph databases. It is assumed that the database administrators will define these views and hence the citable units. MODIS ( Justice, Vermote et al., 1998 ) is an example of a large evolving database of Earth images for which various subcollections have different authorship; and GtoPdb ( Southan, Sharman et al., 2015 ) is a complex curated relational database in which authorship is represented within the database and can be assigned to views determined by the curators.
4.1. Part-of and Reference Annotation
Consider, for simplicity, the case in which the database is static, or that we are only interested in representing citations for one version of the database (we address the more complex case of dynamic databases in the next section). The first observation is that by defining the CUs as views, we immediately obtain a part-of relationship: view V 1 is a part of view V 2 if V 1 can be answered from the result of V 2 . Formally, V 1 is part of V 2 if there is a query Q such that for all possible instances of the database, V 1 ( D ) = Q ( V 2 ( D )).
We have already discussed reference annotations and the information they carry. Among other things, they contain information about where in the cited document the relevant information being cited is to be found. If we look at data citation, this notion of location has much greater importance. For example, the DataCite schema ( DataCite Metadata Working Group, 2016 ; Starr & Gastl, 2011 ) contains the support for the depiction of geospatial data, with properties such as GeoLocation and in particular the subproperty GeoLocationBox , which specifies a bounding box , that is the spatial limits of a box. Most generally we can describe the “location” in the database as a query that extracted the relevant information. This is the approach taken in systems that provide accurate provenance ( Pröll & Rauber, 2013 ). It meshes perfectly with what we are suggesting: The query used to extract the data is a fundamental part of the data citation itself, and the query—or possibly a URL which contains that query—is an essential part of the reference annotation in the citation.
Many approaches can now be defined to decide how to introduce the CU corresponding to data in the citation graph. Here we explore two possibilities, stemming from two of the most used strategies in the research world today. We exemplified these two possible strategies in Figure 3 .

Two examples of possible strategies when citing data. A: Always cite the database. B: Create a view for every new query issued, if it does not already exist, and cite that view.
In Figure 3A we see that a database is represented with a node, DB 1 . A whole database is a citable unit, and every time a paper wants to cite data in that database, it cites the entire database. The reference annotations contain the queries to get the cited data. The paper P 1 presents two citations to DB 1 . Therefore, it has one reference and two reference annotations containing the two different queries being used. P 2 is citing DB 1 only once. The total count of citations to DB 1 is two in this case.
With this solution DB 1 is the only recipient of citations. This means that its number of citations can become very high. On the other hand, it may happen that the rightful authors and curators of the parts of the database being actually cited do not receive any credit for their work.
In Figure 3B we see the strategy adopted by the RDA. Every time a paper cites a data subset extracted through a new query, a citable unit is created in the citation graph; we represent this CU as a view, corresponding to that query. In this case, P 1 is citing DB 1 twice by using two different queries, thus there are two distinct references, corresponding to the two views being cited, V 1 and V 2 . P 2 citing DB 1 with the query Q 3 , generates another view (i.e., V 3 ).
With this solution, new views are created every time it is necessary. This can produce an explosion in the number of nodes in the citation graph, many of which receive only one citation. However, in this way it is possible to cite the exact set of data extracted by the query and to give the credit for the citation to the rightful authors of that data. Moreover, the three views of the example are connected to DB 1 by a “part-of” relationship. This means that DB 1 may inherit all their citations when needed.
We note that to assign only a single CU for the whole database and dispense with the part-of relationship fails when, for example, authorship varies with the part of the database being cited. This is the case with both MODIS and GtoPdb. In this case, we reiterate that it is up to the curators or database administrators to determine the views that define the CUs.
In the case of GtoPdb, both the curators and the contributors agree that the PCU should be the data summary ( Buneman et al., 2020 ) for the most recent view of the database. Unfortunately, the PCU is not determined by the curators but by the system that scans the dedicated journal and creates citation graphs. For a given database, it is the responsibility of the curators or administrators to determine the subsumption relation. Even for conventional publications, we believe that the subsumption relation should be determined by the authors and publishers.
4.2. Dealing with Dynamic Data: Subsumption for Data
Most databases are not static. Unlike documents, they are expected to evolve over time. If versions of a database are released, say, every year, it might be appropriate to treat each version as a new CU. On the other hand, as we discussed, a database in the Citation Graph can present a hierarchy of CUs connected among them through the part-of property. Even though a database may change rapidly, the result of a view, part-of a database, may remain unchanged. The lower a CU is in the part-of hierarchy, the less frequently it will change. Also, even if a part-of CU does change, we may want to treat it as a new version of the previous CU rather than an entirely unrelated new CU, just as we treat an extended or improved version of a paper.
When is it necessary to introduce a new CU representing a view?
If a new CU has been introduced, when can it be considered a new version of the previous CU or an entirely new entity?
If a new CU has been introduced, how do we connect older CUs with the new ones and still keep track of their citation counts?
The answer to the first two questions can only be given by the database administrators. Every time a new version of the database is released, the administrators go through the different CUs that compose the part-of hierarchy of the database and decide which ones need a new version. Recall that subsumption was needed to transfer credit, in the case of papers, from one CU to another: the primary CU (PCU) (i.e., the root of the part-of hierarchy). The same can be done with data.
As we have defined CUs by views when the database changes, we only need to consider creating a new CU if the view changes. More precisely if D and D ′ are successive versions of the database and V is a view, if V ( D ) = V ( D ′) the reference for V ( D ) needs no change, and no new CU is necessary. However if V ( D ) ≠ V ( D ′), we may want to create a new CU .
Once it has been decided that a new CU needs to be created, it is necessary to determine whether the CU associated with V ( D ′) is a new version of the CU for V ( D ), or whether it is, instead, an entirely new CU. The model we propose accommodates both the possibilities; again, this is something that the database administrators or curators can decide. If the content is different in the sense that there is some kind of structural change, then an entirely different CU may be appropriate. Moreover, if the authorship changes, then a different CU may be desirable, as the two versions of the same CU are typically expected to have the same authorship. These are only two examples of reasons why the DBAs may decide to consider the new CU a new, independent, entity.
On the other hand, normally the change will be such that we want the CUs associated with V ( D ) and V ( D ′) to be versions of each other, and the PCU can now become the later version V ( D ′). This preserves the accuracy of the references and allows credit to accumulate on the latest version of the view.
In this second case, it is possible to connect the CU representing V ( D ) to the one representing V ( D ′) through the subsumption relationship. This new relationship has precedence over the part-of relationship, and thus new citations to the older version will be propagated to the new CU, and not upward to the older hierarchy.
Because citation graphs are currently unsuited for representing databases as first-class citizens, we have proposed how to instead extend them to represent data citation in the citation graph. Among other things, this allows us to capture the many citations given to databases and to give credit to the relevant authors or contributors. The new model that we propose is based on a few adjustments, and builds on emerging practices in the world of data citation. Above all, it has the goal of enabling easy adoption, as it is proposed as an extensions of existing models without requiring drastic changes. We argue that, with these extensions of the current model for citation graphs, we can fully achieve the goal of enabling data citation without jeopardizing the existing infrastructures.
The main limitation of existing models on which we have focused are the lack of context on citations between citable units; the inability to deal with different versions of the same CU; and consequently the inability of introduce data, data evolution, and data citation (down to citable portion of a data set) in the citation graph. We showed how, by solving the first two problems through the introduction of reference annotations and the subsumption property, we are also able to model data citation appropriately in the citation graph.
Unlike traditional scholarly publications, databases present a greater range of granularities and are subject to more frequent change. Concerning the granularity of data, although it is possible to consider various scenarios, we work with two main cases: either only the whole database is treated as a node, or each time a new query is issued, a new node is added to the graph, connected to the whole database through a part-of relationship.
The first solution is similar to what already happens with papers in data journals. In this case, the whole database is represented through a single CU (i.e., one node in the graph). Every time a paper cites data in the database, the citation goes to the database. Information such as the query and the rightful authors of the citation may be inserted in the reference annotation of the citation. This solution is simple, but gives all the citations to the whole database, thus without explicit recognition for the rightful curators of the cited data. Therefore, more computations are necessary to obtain the citation counts of the single queries and the corresponding authors.
With the second solution, which follows the RDA specifications ( Rauber, Asmi et al., 2015 ), every time a new query is issued to the database, a new CU (hence, a new node) is created. In this case, the graph represents explicitly what is cited, and thus the rightful owners receive their citations without further computations. However, this solution may result in an explosion of nodes. To mitigate this problem different techniques could be deployed. For example, it could be possible to use algorithms of query containment to decide when a query behind a citation can be answered from a CU already deployed. In this case, that CU could receive that citation, instead of creating a new node. Of course, query containment is, in general, an NP-hard problem, and it could become computationally prohibitive to exploit this solution, in particular in situations where many nodes have already been created. Alternatively, the system could present to the interested user a series of precomputed queries, corresponding to already instantiated CUs, which may suit their citing needs. In this way, the system already knows to which node to assign the citation.
We also observe that it could be possible to extend the proposed data model where, instead of nodes presenting the metadata of the papers, the CUs are represented using or including the annotated full text of a paper. In this way, annotations on the paper can be used to keep track of different types of information, such as references and reference annotations. Although this solution has greater expressive power, it also increases the size and complexity of the model. As already discussed, the model proposed in this paper has the advantage of being easy to implement on top of already existing systems. A new model, considering the whole annotated text of a paper presents new implementation challenges, and thus requires the creation of a new application from scratch.
It is important also to note how, as of today, there are many challenges to the implementation and proper operation of data citation in general. Often the RDA guidelines for dynamic data citation are not implemented by many databases; it is often difficult to automatically produce context and thus reference annotations that are machine readable; and there are also many bad practices among researchers, such as that of depositing PDFs, images, and tables of their papers in data repositories, calling them research data. Although there are still many hindrances to the correct implementation of data citation, the research community has still showed a great desire for the implementation of common techniques and best practices for the correct application of these guidelines. Databases such as Eagle-i 15 already provide data citation snippets, whereas others, like GtoPdb, automatically produce PDFs of their pages to allow an easier citation of their data in form of CUs. Thus, it is our conviction that as data citation gets more traction and is implemented appropriately, it would be crucial to account for it and integrate such information in the common citation graph. In particular, a model such as the one we propose in this paper will allow a better and fairer implementation of data citation to be achieved, and will also benefit all researchers and become more and more needed as we transition toward the fourth paradigm of science. The more we learn about the current limits of data citation and how to address them, the faster we will come to the final goal of a correct system for citing data.
Considering new possible research problems, we note that the citation graph in fact is, among other things, a historical record , that is, a record of how researchers interacted with information and other works to build their expertise and new knowledge. Given this interpretation, then the graph should not be rewritable, that is, it should not be possible to rewrite history . Therefore, the graph should be a timestamped “append-only” structure in a way similar to the distributed ledgers. Thus, it should only be possible to insert data in it without the possibility to overwrite or modify already existing information.
Among others, these requirements are necessary for the computation of impact factors ( Garfield, 2006 ) where it is necessary to know the number of citations received by a journal in the past 2 years. It is therefore mandatory that this information is not modified over time. This is true also for other types of statistics that researchers may be interested in.
In our examples, we have taken care to timestamp every element of information to make this possible. The timestamps in particular indicate the moments the events “occurred” (e.g., when a citation happened), not when they were inserted in the graph. However, there are several issues concerning the semantics and representation of temporal information in the citation graph that require further investigation.
If this property is correctly implemented, it should enable one to perform different types of query on the graph. That is, past versions of the database should be accessible for accurate provenance. Ideally, given the state of the graph in the present, it should be possible to rebuild a previous state at any given time in the past. We call this property history preservation .
Several databases have this property. Weather data and geospatial data are generally accumulative ( Justice et al., 1998 ). Blockchains are also based on the idea that once added, a block cannot be removed or modified, to guarantee the preservation of the history of the transactions.
On the other hand, curated databases are not, in general, history preserving, in the sense that they are updated and change with time. This is particularly problematic for data citation because one of its desiderata is that a citation should always allow retrieving or at least knowing what was cited ( Buneman, 2006 ). Therefore, we see the correct extension and implementation of history preservation as an important future challenge to be tackled in the implementation of a data-aware citation graph.
6.1. Databases in Relation to Data Citation
As we mentioned above, there are three main categories of databases that can be cited: static databases; evolving databases; and curated databases. As a reasonable generalization, the problem of data citation is easily solved for the first category, as many systems and practices have been developed for static databases. In this case, databases are treated as they were traditional publications because they are never updated, the list of authors does not change, and even though only a portion of the database is cited, the citation goes to the whole database. In this case, when we consider the citation graph, we have one single node representing a database receiving all the citations from papers and data.
For the other two cases, data citation remains problematic. One relevant open issue is the citation of data subsets generated on the fly by issuing general queries to the database. In this case, the main problems are how to guarantee the persistence and accessibility of the data in the cited form and automatically provide a complete and correct textual reference for the cited data.
The first problem is tackled by the RDA 16 . The RDA is a community-driven initiative launched in 2013 by different commissions, including the European Commission and the US government’s National Science Foundation. Its goal is to build the social and technical infrastructures to enable open sharing and reuse of data. The RDA “Working Group on Data Citation: Making Dynamic Data Citable” (WGDC) 17 ( Rauber, Ari et al., 2016 ) has been working in recent years on large, dynamic, and changing data sets. Although the WGDC first focused on RDBMs as the first forms of pilot solutions, many other types of databases followed (XML, CSV, files, Git repositories, distributed databases such as VAMDC ( Zwölf, Moreau et al., 2019 ), and multidimensional data cubes such as NetCDF/CCCA ( Schubert, 2017 )). The working group has finished the development of its guidelines, and has now moved on into an adoption phase.
In particular, among the goals of the RDA WGDC ( Rauber et al., 2015 ), there is the identification and citation of arbitrary views of data. As potential solution, WGDC recommends an identification method based on assigning PIDs to queries, that are then used as proxies for the data subset to be cited. The access to a data subset is enabled by reissuing the stored query and a citation is associated with the PID of the query identifying the data ( Rauber et al., 2016 ). A PID is an identifier meant to uniquely and persistently (i.e., continually during the course of time) identify an object such as a publication, data set, or person, usually in the context of digital objects that are accessible over the internet. Considering the citation graph, this method based on PID adds a new citable unit every time a new query is cited and requires to check query equivalence (and/or containment) to avoid the creation of a new citable unit for an already cited query.
The second aspect is characterized as a computational problem ( Buneman et al., 2016 ) and some solutions based on “query rewriting using views” ( Davidson, Buneman et al., 2017 ) have been proposed, targeting general queries citations for relational databases ( Alawini, Davidson et al., 2017b ; Wu, Alawini et al., 2018 ; Wu, Alawini et al., 2019 ) and graph databases ( Alawini, Chen et al., 2017a ).
Overall, most approaches do not consider the evolution of data and the fact that databases are not monolithic objects. When those features are considered, some of the existing models propose the trivial solution of treating databases and views as standalone objects. In our model, instead, we explicitly model citable units and their subsumption relationships, which allow the appropriate distribution of credit.
6.2. Available Citation Graphs
The citation graph, or citation network, as a model of a graph where the vertices represent academic papers, has long been in use in the literature ( Price, 1965 ) and has evolved considerably. There are different implementations of citation graphs, which favor certain aspects of the information regarding publications, citations, and authors, depending on the considered task. Some of them are provided explicitly for navigational purposes (e.g., the Open Academic Graph (OAG)). Others, instead, are the backbone of services allowing search and exploration of scholarly works; these are the Microsoft Academic Graph (MAG), Google Scholar, PubMed, Web of Science, Scopus, and Semantic Scholar.
The Microsoft Academic Graph (MAG) ( Färber, 2019 ; Wang, Shen et al., 2019 ) is the backbone of the Microsoft Academic Service (MAS), and its nodes represent five different entities: field of study, author, institution, paper, venue, and event. An RDF version of MAG, called Microsoft Academic Knowledge Graph 18 (MAKG) is also available and connected to the Linked Open Data cloud.
The Open Academic Graph (OAG) 19 is an open-source citation graph generated from the linking of two other large academic graphs: MAG and ArnetMiner (or AMiner) ( Wan, Zhang et al., 2019 ) (a free online service used to index, search, and mine big scientific data), designed to search and perform data mining against academic publications available on the Internet. This graph contains entities similar to those of MAG, and it can be used as a unified sizable academic graph for the study of citation networks, paper content, and the integration of multiple academic graphs with different fields and information.
The OpenAIRE Research Graph ( Manghi et al., 2019 ) is the implementation of a fully fledged Open Science Graph. It is a collection of metadata and links connecting research entities, including articles, data sets, software, etc., together with other elements such as organizations, funders, funding streams, projects, research communities, and data sources 20 . The graph today contains around 110 million publications, 10 million data sets, 180,000 software research products, and 7 million other products with 480 million links between them. The aim of the OpenAIRE RG is to bring discovery, monitoring, and assessment of science into the hands of the scientific community ( Fava, 2020 ).
The PID Graph ( Fenner, 2020 ; Fenner & Aryani, 2019 ) is another example of implementation of a citation graph based around the concept of PID (Persistent IDentifier). The PID Graph targets citations aggregation: for all versions of a data set or software source code; for all data sets hosted in a particular repository, funded by a particular funder, or aggregated by a particular researcher; and for a research object, such as a publication or the data used in the paper, together with the software and samples used to create the data set. The PID graph adopts the outputs of the RDA WGDC. One peculiarity of the PID graph is that it includes not only metadata about connections but also metadata about the resources and implicit relations about resources identified by the PIDs. This enables queries based on these metadata, making them more expressive.
Google Scholar, PubMed, Web of Science, and Scopus are all relevant services providing citation graphs, but their data is not directly accessible as a graph.
Google Scholar is an open general-purpose graph focusing on traditional publications and covering multiple languages and publication venues. PubMed, instead, focuses on medicine and biomedical sciences ( Roberts, 2001 ). It covers medical bibliography from 1949 until today, with abstracts, review articles, and free full-text articles. Web of Science (WoS) provides subscription-based access to multiple databases with comprehensive citation data for many different academic disciplines ( Falagas, Pitsouni et al., 2008 ). Finally, Scopus is Elsevier’s abstract and indexing (closed) database featuring open access titles, indexes of web pages and patents, and links to both citing and cited documents ( Burnham, 2006 ). Although PubMed is an important resource for clinicians and researchers, Scopus covers a wider journal range, offering also the capability for citation analysis, although limited with respect to WoS, which covers articles published before 1995. Google Scholar, on the other hand, presents all the pros and cons of a web search engine: It can help in the retrieval also of oblique information, but it may present inadequate and less often updated citation information ( Falagas et al., 2008 ).
Semantic Scholar is a project developed at the Allen Institute for Artificial Intelligence and is an AI-backed search engine for scientific journal articles. It uses a combination of machine learning, natural language processing, and machine vision to produce a semantic analysis of the papers of the network and to extract figures, entities, and venues from the documents. It is designed to highlight the most important and influential articles and to identify the connections between them ( Fricke, 2018 ).
As we can see, many of these graphs and systems could work as good starting points for the implementation of the proposed model. MAG and OAG already present the context, which can be used as reference annotation, but lack the ability to accurately cite data and manage their versions. On the other hand, the OpenAIRE graph is able to deal with granularity and different versions, but it still lacks the possibility to cite its data with reference annotations; thus de facto it is still unable to deal with data citations. Nonetheless, many of the systems implemented are close to the proposed model, and usually they lack one aspect (like the versioning of the data or the presence of context). Therefore, we believe that a viable way forward would be to implement the approach we propose on top of the already existing infrastructures.
Applications of the citation graphs are disparate. Some examples include prediction of user queries over the graph; recommendation systems for the generation of suggestions leveraging the relationships across the different types of entities; exploration of papers, researchers, affiliations, and other entities; data integration; data analysis; and knowledge discovery of scholarly data through expert finding, geographic search, trend analysis, review recommendation, association search, course search, academic performance evaluation, and topic modeling ( Wan et al., 2019 ).
Given the vital role of citation graphs and data citation, we argue that it is of crucial importance that existing citation graphs be extended with the appropriate tools to model data citation in various forms. Most of the existing citation graph do not expose their internal data model. Nonetheless, we can see they focus on the same core assumption that citable objects are atomic elements with no citable portions and where evolution through time is not considered. Hence, none of the models above tackle explicitly and directly the issues linked to the task of modeling databases and subsets of databases, as well as the evolution of citable elements through time, which is instead the goal of this work.
Starting from a basic model of the citation graph in which the nodes are the papers, and the edges are the citations between them, we highlighted three limitations of this model. They are the lack of context for citations, that is, information about the how and why the citation is used along with which part of the referenced object is used; the absence of a unified strategy of management of the versions of the papers in the graph; and the difficulty of representing citations to databases and data generated by queries in the graph.
To deal with these limitations, we proposed an implementation-agnostic model that includes reference annotations. These annotations contain the context of a citation (e.g., the page numbers of the citation, the query issued to obtain the data, or the considered bounding box).
We also discussed the subsumption property, which is used when a new version of a paper or a database is introduced in the graph. This property indicates that the new version “takes the place” of the previous one for the purpose of assigning credit. The old citations can be inherited from the new version or, depending on the context, such as situations where authors have changed, different policies can be put in place.
Although we have used subsumption partly to deal with the evolution of citable units within databases, we believe there is much more to be said about evolution in databases and in the citation graph itself. We believe that all scientific databases should support “time travel”: it should be possible to ask queries on some previous state of the database as easily as one asks queries on the current state. For many databases, especially “source data,” it is important to support longitudinal queries, and this is true of the citation graph itself.
We have dealt with citations to databases, but what about citations from databases? If, as happens in many curated databases, conventional citations are included within the database, then there should be few problems, but what happens when a part of one database is created by a query from another database? How is the citation represented; and how is it included in the citation graph?
Finally, once we have properly supported databases within the citation graph, what kinds of bibliometric measures are possible? We have, for example, h -indexes and impact factors for conventional publications. How can we appropriately measure the impact of databases?
We note that there is currently marginal interest to cite software and code, even though interesting initiatives, such as the FORCE 11 working group 21 , have been taking place and research groups are working on the topic ( Alliez, Di Cosmo et al., 2020 ; Katz, Niemeyer et al., 2016 ; Katz, Bouquin et al., 2019 ). This task presents a new set of problems, in particular regarding authorship, because code is often copied or adapted from other repositories, passing from hand to hand, undergoing modifications. The characteristics of the life cycle of software open a whole new set of problems and research questions about who is the righteous author of that piece of cited code and who should receive credit from the citation.
The authors would like to thank the reviewers for their detailed comments and suggestions.
Following the CRediT guidelines 22 , all authors contributed equally to the conceptualization, investigation, methodology, and writing of the paper.
The authors declare that they do not have any competing interest.
The work was partially supported by the ExaMode project, as part of the European Union H2020 program under Grant Agreement No. 825292. Matteo Lissandrini is supported by the European Union H2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No. 838216.
Not applicable.
https://www.microsoft.com/en-us/research/project/microsoft-academic-graph/
https://www.semanticscholar.org/
https://www.aminer.cn/
https://www.ncbi.nlm.nih.gov/pubmed/
https://www.scopus.com/home.uri
https://clarivate.com/webofsciencegroup/solutions/web-of-science/
See https://fairsharing.org/databases/ for a detailed list of curated scientific databases commonly used in research.
https://datasetsearch.research.google.com/
https://www.rd-alliance.org/
https://www.w3.org/ns/oa#Annotation
https://www.mendeley.com/
https://zenodo.org/
Property graphs are an exception because they allow data to be assigned to edges.
https://tinyurl.com/y9clyx8d , retrieved March 16, 2020
https://www.eagle-i.net/
https://www.rd-alliance.org/groups/data-citation-wg.html
https://ma-graph.org
https://aminer.org/open-academic-graph
https://graph.openaire.eu
https://www.force11.org/group/software-citation-working-group
https://casrai.org/credit/
Email alerts
Related articles, affiliations.
- Online ISSN 2641-3337
A product of The MIT Press
Mit press direct.
- About MIT Press Direct
Information
- Accessibility
- For Authors
- For Customers
- For Librarians
- Direct to Open
- Open Access
- Media Inquiries
- Rights and Permissions
- For Advertisers
- About the MIT Press
- The MIT Press Reader
- MIT Press Blog
- Seasonal Catalogs
- MIT Press Home
- Give to the MIT Press
- Direct Service Desk
- Terms of Use
- Privacy Statement
- Crossref Member
- COUNTER Member
- The MIT Press colophon is registered in the U.S. Patent and Trademark Office
This Feature Is Available To Subscribers Only
Sign In or Create an Account
New tool to visualize related articles
- Author By ame5
- Publication date February 3, 2021
- Categories: arXivLabs

Readers will find the feature below an article abstract in the “Related Papers” tab, as shown below. By activating the Connected Papers toggle switch, readers can follow a link to the article’s graph displayed at Connected Papers. Each paper’s graph is created by analyzing tens of thousands of papers for similarity in their citations, and then a small subset of those analyzed are arranged according to their degree of similarity. Each node in the graph represents an article, which has its own set of connected papers.
“Connected Papers started as a weekend side project between friends, to solve our own problems with literature reviews,” said Eddie Smolyansky, co-founder of Connected Papers. “We can’t believe how quickly the scientific community embraced the tool and we’re so excited to be featured on arXiv – a website that we use daily in our own research. With this kind of support, we plan to keep improving Connected Papers and to build more tools for the academic community.”
arXivLabs is a framework enabling innovative collaborations with individuals and organizations to bring innovative tools to the arXiv community, and we welcome new proposals .
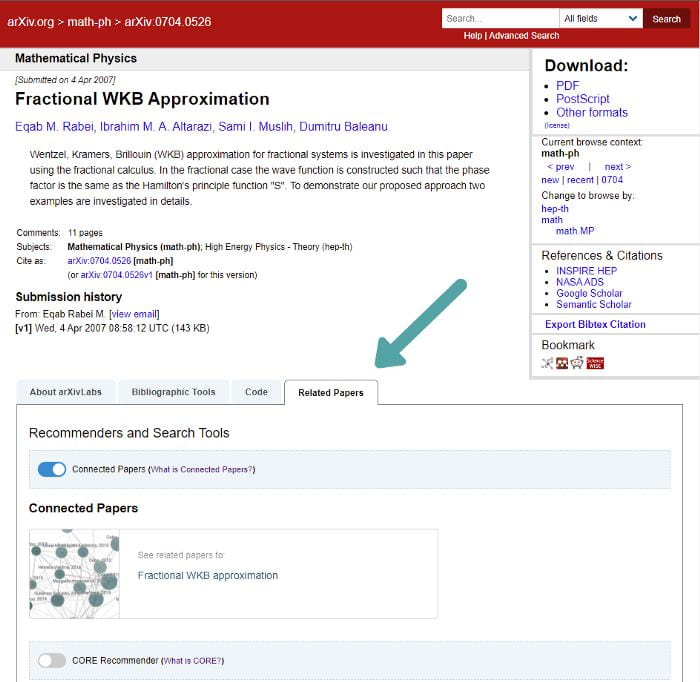
Subscribe By Email
Get every new post delivered right to your inbox.
Your Email Leave this field blank
This form is protected by reCAPTCHA and the Google Privacy Policy and Terms of Service apply.

Connected Papers: a visual tool for academic research
Anne-Laure Le Cunff
I’m obsessed with thinking in maps : discovering and creating connections between ideas, adding nodes to a knowledge graph, finding patterns across distant areas of knowledge. However, the traditional way of exploring connections between research papers is fairly tedious: read the paper, scan the references, search for any relevant title, rinse and repeat. Connected Papers aims to shake things up.
Connected Papers is a tool for thought to help researchers and applied scientists find and explore papers relevant to their field of work in a visual way. You enter an origin paper, and they generate a graph. To achieve this, they analyse about 50,000 research papers, and select the ones with the strongest connections to the origin paper.
Created by Alex Tarnavsky, Eitan Eddie Smolyansky, and Itay Knaan Harpaz from Israel, Connected Papers started as a weekend side project. But when the three friends realised how useful it was in their own research, and how their friends and colleagues kept on asking to use it, they decided to release the tool for the public.
Some of the benefits of Connected Papers include:
- Getting a visual overview of a field of research. You will be able to see at a glance which papers are most popular in the field, as well as the various dynamics between areas of studies.
- Making sure you haven’t missed a key paper. This is especially useful in fields that constantly produce a large volume of new papers.
- Exploring relevant papers in a bi-directional manner. Connected Papers lets you discover the most important prior and derivative work in your area of interest.
The tool is currently completely free, and the three co-founders keep on adding new features to make it even more useful. If you want to give it a try, follow these instructions.
1. Enter an origin paper
On the home page, enter one of the options to identify your origin paper. You can use a DOI, the paper’s title, or the paper’s URL from arXiv, PubMed, or Semantic Scholar. Then click on “Build a graph. For this tutorial, I used this paper , which can read more about here .
2. Read the graph
On the next page, you will be greeted by three panels. We’ll discuss the other panels later, but for now, let’s focus on the graph. Each node is a research paper related to the origin paper. Rather than a basic citation tree, the papers are arranged according to their similarity.
The size of a node represents the number of citations. The color of a node represents the publishing year—lighter is older. You will notice that highly similar papers have stronger connecting lines and tend to cluster together.
3. Explore the graph
You can scroll through papers in the left panel. Whenever you click on a paper there, it will be highlighted on the graph. You can also navigate the graph by clicking on specific nodes. Both options will update the right-side panel with more information about the selected paper.
Two buttons in the top left corner allow you to explore papers that are not included in the graph, but probably relevant to your topic of choice.
- Prior works. These are research papers that were most commonly cited by the papers included in the graph. It usually means that they are important seminal works for this field. Selecting a prior work will highlight all graph papers referencing it in the left-side panel, and selecting a graph paper will highlight all referenced prior work.
- Derivative works. These are research papers that cited many of the graph papers. It probably means they are either recent relevant works or surveys of the field. Similar to prior works, “selecting a derivative work will highlight all graph papers cited by it, and selecting a graph paper will highlight all derivative works citing it.”
If you find a paper particularly promising, you can click on “paper details” to open the link to the paper in a new window, or on “build a graph” to create a new graph based on this origin paper. Building the new graph can sometimes take a few seconds, but there will be a progress bar so you know how long to wait.
All of your graphs can be found in the top right corner of the tool, under “my graphs”.
Connected Papers is incredibly well designed, easy to use, and most importantly very helpful in exploring research paths of influence. I highly recommend giving it a try to build your mental atlas .
Update: Connected Papers is now supported on mobile browsers !
Join 80,000 mindful makers!
Maker Mind is a weekly newsletter with science-based insights on creativity, mindful productivity, better thinking and lifelong learning.
One email a week, no spam, ever. See our Privacy policy .
Don’t work more. Work mindfully.
Ness Labs provides content, coaching, courses and community to help makers put their minds at work. Apply evidence-based strategies to your daily life, discover the latest in neuroscience research, and connect with fellow mindful makers.
Ness Labs © 2022. All rights reserved .
Citation Tree
Enter a DOI to display the citation tree
Grasp a fundamental understanding of a field from a couple of core papers
Enter the DOI of your article of choice below to visualize its scientific reference environment:
Spot core papers describing a field as shown below:
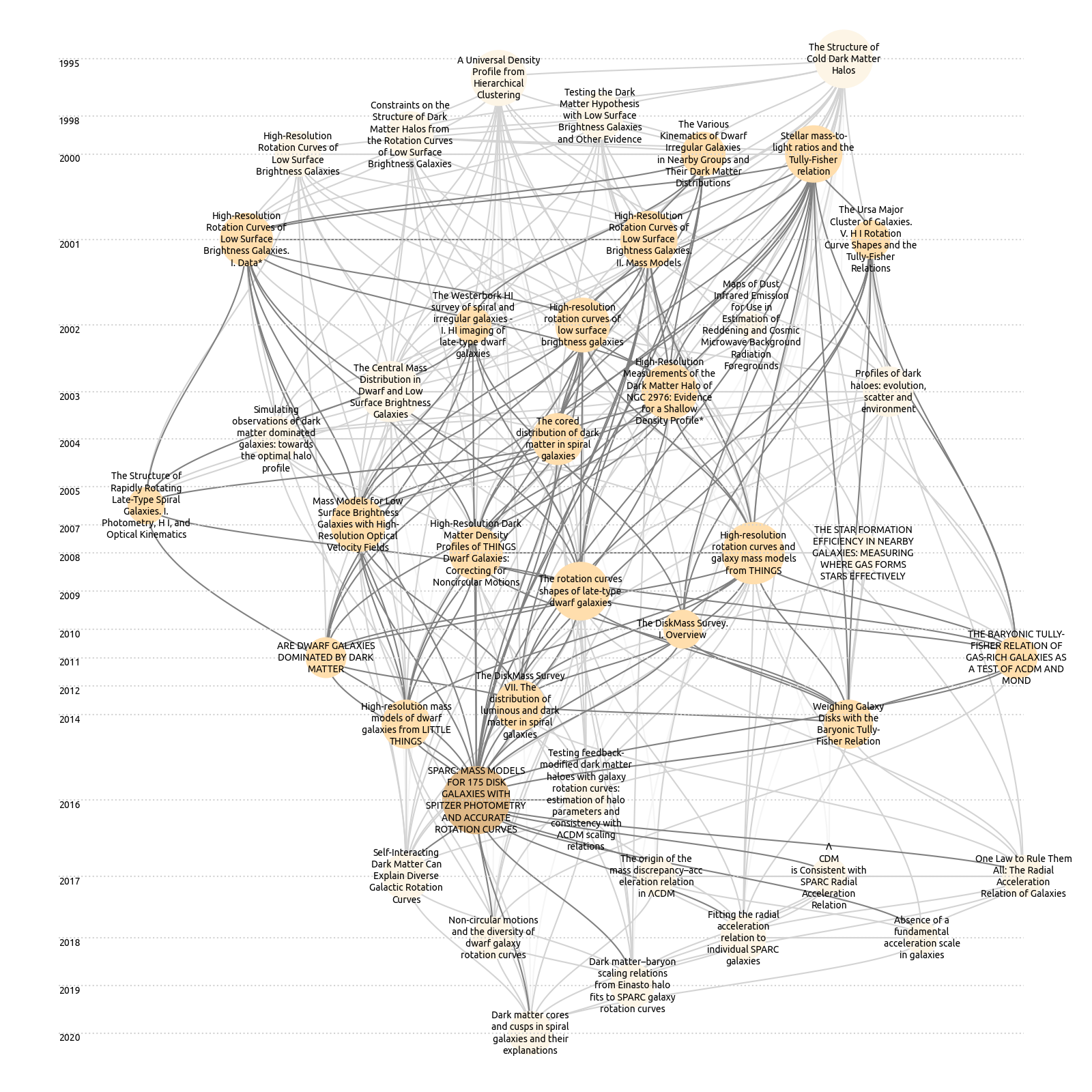
SPARC: Mass models for 175 disk galaxies with Spitzer phtometry and accurate rotation curves.
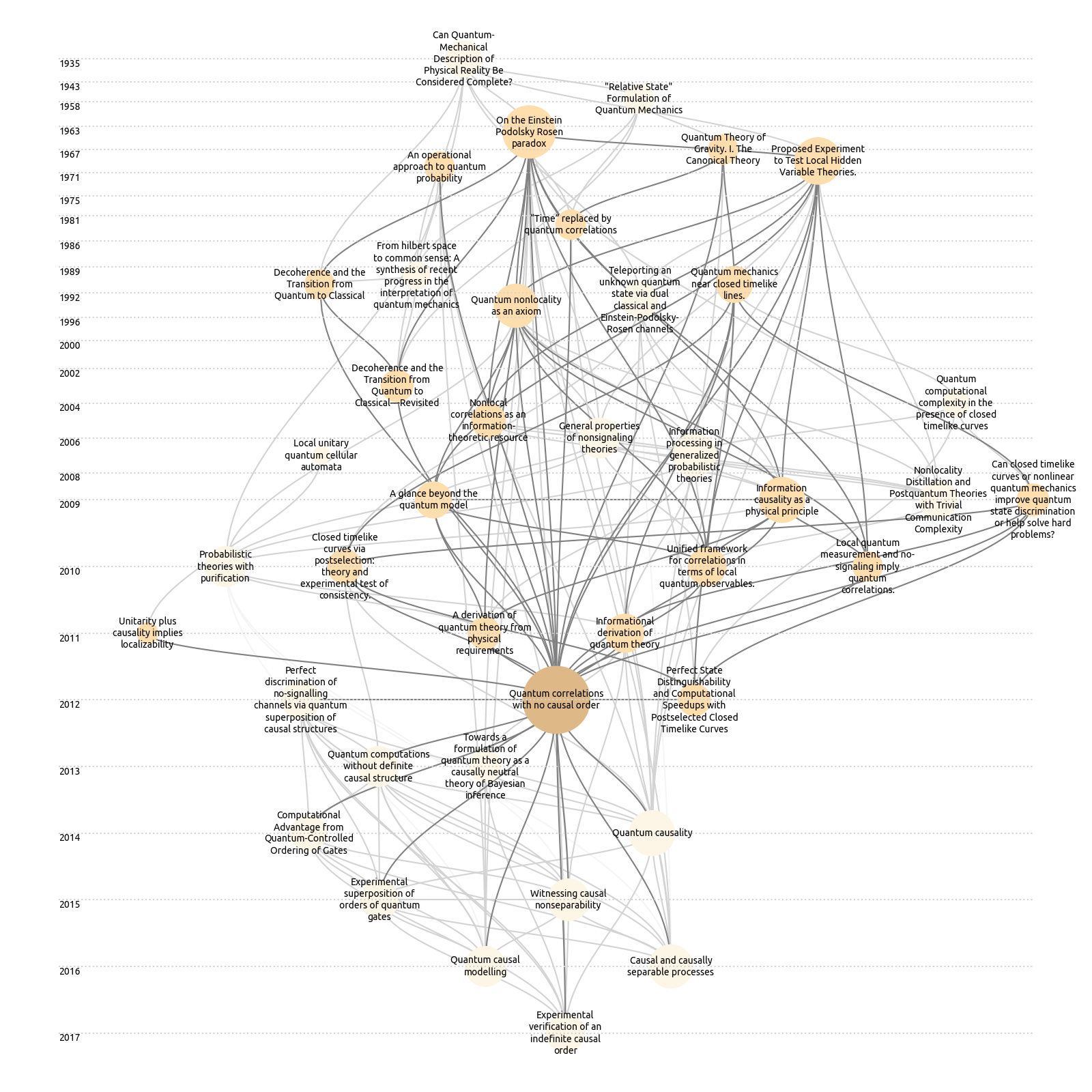
Quantum correlations with no causal order.
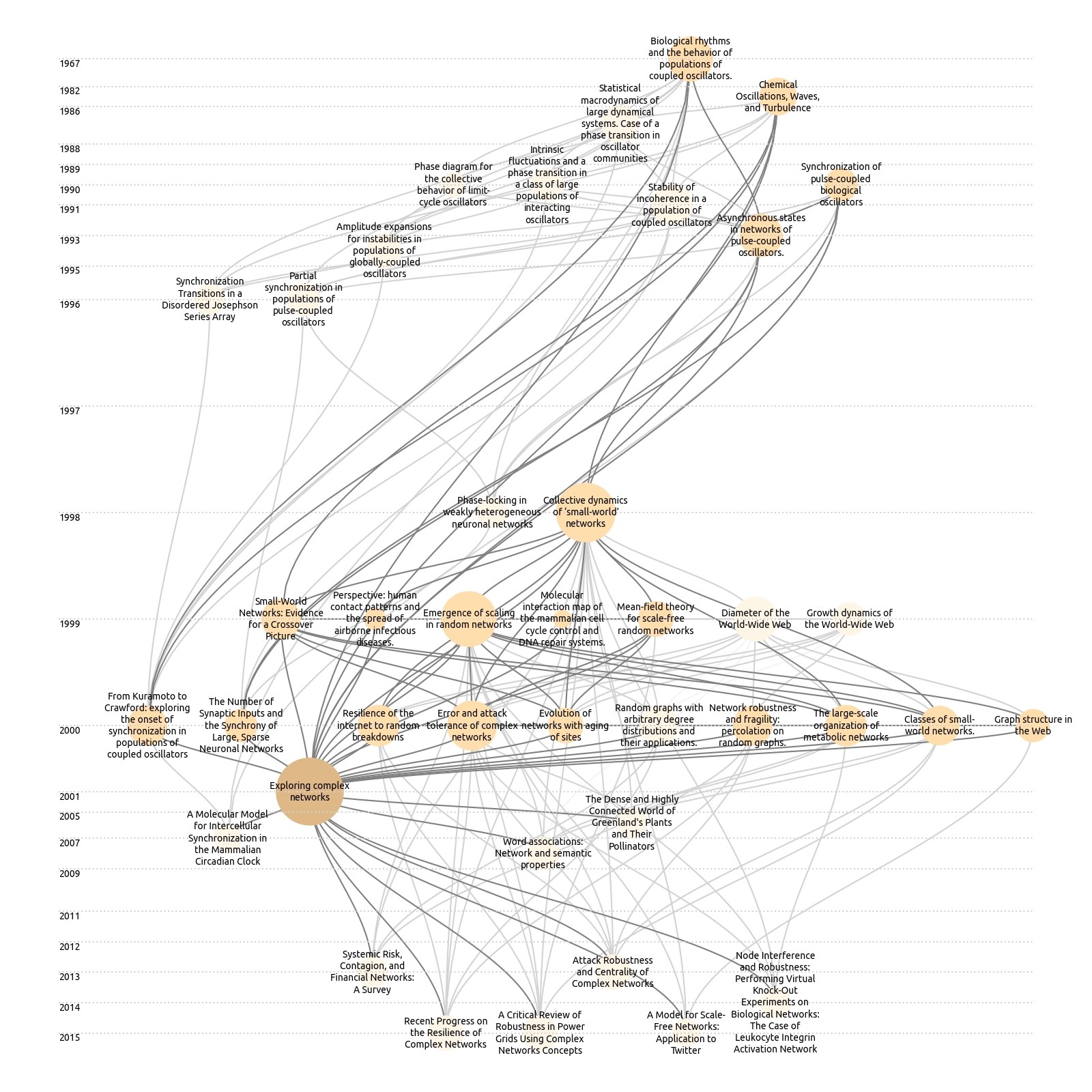
Exploring complex networks.
Ideas originate in a couple of Important articles. Then flourish in different flavors amongst scientific literature. Now get a quick visualization of citation trees. This is useful to have an overview of a new field, to spot important papers and accelerate your bibliography. We use data from Crossref and Semantic Scholar .
Welcome to Citegraph
Citegraph is an open-source online visualizer of 5+ million papers, 4+ million authors, and various relationships. In total, Citegraph has 9.4 million vertices and 294 million edges. At the moment, Citegraph only has computer science bibliography.
Paper ---CITES--> Paper
Citegraph contains 32+ million paper citation edges.
Fun fact: Distinctive Image Features from Scale-Invariant Keypoints is the most cited paper.
Author ---WRITES--> Paper
Citegraph contains 16+ million authorship edges.
Fun fact: H. V. POOR is the most productive researcher - he has authored more than 1.6k papers!
Author ---REFERS--> Author
Citegraph contains 224+ million author citation edges.
Fun fact: Geoffrey Hinton is the most-cited person - more than 66k people have cited his work at least once!
Author ---COLLABORATES with--> Author
Citegraph contains 19+ million author collaboration edges.
Fun fact: Radu Timofte has the most collaborators - more than 2k people have coauthored at least one paper with him!
Generate accurate APA citations for free
- Knowledge Base
- Citing tables and figures from other sources in APA Style
Citing Tables and Figures in APA Style | Format & Examples
Published on November 6, 2020 by Jack Caulfield . Revised on December 27, 2023.
When you reprint or adapt a table or figure from another source, the source should be acknowledged in an in-text citation and in your reference list . Follow the format for the source type you took the table or figure from.
You also have to include a copyright statement in a note beneath the table or figure. The example below shows how to cite a figure from a journal article .
Table of contents
Citing tables and figures, including a copyright note, examples from different source types, frequently asked questions about apa style citations.
Tables and figures taken from other sources are numbered and presented in the same format as your other tables and figures . Refer to them as Table 1, Figure 3, etc., but include an in-text citation after you mention them to acknowledge the source.
You should also include the source in the reference list. Follow the standard format for the source type you took the table or figure from.
Prevent plagiarism. Run a free check.
As well as a citation and reference, when you reproduce a table or figure in your own work, you also need to acknowledge the source in a note directly below it.
The image below shows an example of a table with a copyright note.
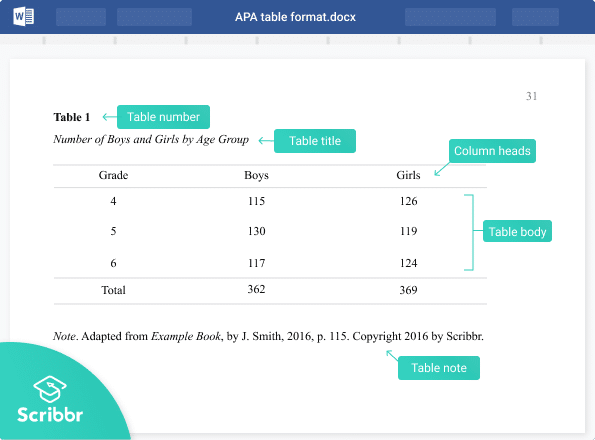
If you’ve reproduced a table or figure exactly, start the note with “From …” If you’ve adapted it in some way for your own purposes (e.g. incorporating part of a table or figure into a new table or figure in your paper), write “Adapted from …”
This is followed by information about the source (title, author, year, publisher, and location), and then copyright information at the end.
Types of copyright and permission
A source will either be under standard copyright, under a Creative Commons license, or in the public domain. You need to state which of these is the case.
Under standard copyright, you sometimes also need permission from the publisher to reprint or adapt materials. If you sought and obtained permission, mention this at the end of the note.
Look for information on copyright and permissions from the publisher. If you’re having trouble finding this information, consult your supervisor for advice.
- From a journal article
- From a website
- From a book
Copyright information can usually be found wherever the table or figure was published. For example, for a diagram in a journal article , look on the journal’s website or the database where you found the article. Images found on sites like Flickr are listed with clear copyright information.
If you find that permission is required to reproduce the material, be sure to contact the author or publisher and ask for it.
APA doesn’t require you to include a list of tables or a list of figures . However, it is advisable to do so if your text is long enough to feature a table of contents and it includes a lot of tables and/or figures .
A list of tables and list of figures appear (in that order) after your table of contents, and are presented in a similar way.
If you adapt or reproduce a table or figure from another source, you should include that source in your APA reference list . You should also include copyright information in the note for the table or figure, and include an APA in-text citation when you refer to it.
Tables and figures you created yourself, based on your own data, are not included in the reference list.
In most styles, the title page is used purely to provide information and doesn’t include any images. Ask your supervisor if you are allowed to include an image on the title page before doing so. If you do decide to include one, make sure to check whether you need permission from the creator of the image.
Include a note directly beneath the image acknowledging where it comes from, beginning with the word “ Note .” (italicized and followed by a period). Include a citation and copyright attribution . Don’t title, number, or label the image as a figure , since it doesn’t appear in your main text.
Cite this Scribbr article
If you want to cite this source, you can copy and paste the citation or click the “Cite this Scribbr article” button to automatically add the citation to our free Citation Generator.
Caulfield, J. (2023, December 27). Citing Tables and Figures in APA Style | Format & Examples. Scribbr. Retrieved April 2, 2024, from https://www.scribbr.com/apa-examples/citing-tables-figures/
Is this article helpful?

Jack Caulfield
Other students also liked, how to format tables and figures in apa style, how to cite an image in apa style, setting up the apa reference page | formatting & references (examples), scribbr apa citation checker.
An innovative new tool that checks your APA citations with AI software. Say goodbye to inaccurate citations!

APA Citation Guide (7th edition) : Images, Charts, Graphs, Maps & Tables
- What Kind of Source Is This?
- Advertisements
- Books & eBooks
- Book Reviews
- Class Handouts, Presentations, and Readings
- Encyclopedias & Dictionaries
- Government Documents
- Images, Charts, Graphs, Maps & Tables
- Journal Articles
- Magazine Articles
- Newspaper Articles
- Personal Communication (Interviews, Emails)
- Social Media
- Videos & DVDs
- Paraphrasing
- Works Cited in Another Source
- No Author, No Date etc.
- Sample Paper, Reference List & Annotated Bibliography
- Powerpoint Presentations

On This Page
Image reproduced from a magazine or journal, image reproduced from a website.
Reproducing Images, Charts, Tables & Graphs
Reproducing happens when you copy or recreate an image, table, graph or chart that is not your original creation. If you reproduce one of these works in your assignment, you must create a note underneath the image, chart, table or graph to show where you found it. You do not include this information in a Reference list.
Citing Information From an Image, Chart, Table or Graph
If you refer to information from an image, chart, table or graph, but do not reproduce it in your paper, create a citation both in-text and on your Reference list.
If the information is part of another format, for example a book, magazine article, encyclopedia, etc., cite the work it came from. For example if information came from a table in an article in National Geographic magazine, you would cite the entire article.
If you are only making a passing reference to a well known image, you would not have to cite it, e.g. describing someone as having a Mona Lisa smile.
Figure Numbers
Each image you reproduce should be assigned a figure number, starting with number 1 for the first image used in the assignment.
Images may not have a set title. If this is the case give a description of the image where you would normally put the title.
Copyright Information
When reproducing images, include copyright information in the citation if it is given, including the year and the copyright holder. Copyright information on a website may often be found at the bottom of the home page.
Note: Applies to Graphs, Charts, Drawings, Maps, Tables and Photographs
Figure X . Description of the image or title of the image. From "Title of Article," by Article Author's First Initial. Second Initial. Last Name, year, day, (for a magazine) or year (for a journal), Title of Magazine or Journal, volume number, page(s). Copyright year by name of copyright holder.
Note : Information about the image is placed directly below the image in your assignment. If the image has been changed, use "Adapted from" instead of "From" before the source information.
Figure 1 . Man exercising. Adapted from "Yoga: Stretching Out," by A. N. Green, and L. O. Brown, 2006, May 8, Sports Digest, 15 , p. 22. Copyright 2006 by Sports Digest Inc.
Note: Applies to Graphs, Charts, Drawings, Tables and Photographs
Figure x. Description of the image or image title if given. Adapted from "Title of web page," by Author/Creator's First Initial. Second Initial. Last Name if given, publication date if given, Title of Website . Retrieved Month, day, year that you last viewed the website, from url. Copyright date by Name of Copyright Holder.
Note : Information about the image is placed directly below the image in your assignment. If the image has not been changed but simply reproduced use "From" instead of "Adapted from" before the source information.
Figure 2 . Table of symbols. Adapted from Case One Study Results by G. A. Black, 2006, Strong Online. https://www.strongonline/ casestudies/one.html. Copyright 2010 by G.L. Strong Ltd.
- << Previous: Government Documents
- Next: Journal Articles >>
- Last Updated: Jan 5, 2024 2:56 PM
- URL: https://columbiacollege-ca.libguides.com/apa
- AUT Library
- Library Guides
- Referencing styles and applications
APA 7th Referencing Style Guide
- Figures (graphs and images)
- Referencing & APA style
- In-text citation
- Elements of a reference
- Format & examples of a reference list
- Conferences
- Reports & grey literature
General guidelines
From a book, from an article, from a library database, from a website, citing your own work.
- Theses and dissertations
- Audio works
- Films, TV & video
- Visual works
- Computer software, games & apps
- Lecture notes & Intranet resources
- Legal resources
- Personal communications
- PowerPoint slides
- Social media
- Specific health examples
- Standards & patents
- Websites & webpages
- Footnotes and appendices
- Frequently asked questions
A figure may be a chart, a graph, a photograph, a drawing, or any other illustration or nontextual depiction. Any type of illustration or image other than a table is referred to as a figure.
Figure Components
- Number: The figure number (e.g., Figure 1 ) appears above the figure in bold (no period finishing).
- Title: The figure title appears one double-spaced line below the figure number in Italic Title Case (no period finishing).
- Image: The image portion of the figure is the chart, graph, photograph, drawing, or illustration itself.
- Legend: A figure legend, or key, if present, should be positioned within the borders of the figure and explain any symbols used in the figure image.
- Note: A note may appear below the figure to describe contents of the figure that cannot be understood from the figure title, image, and/or legend alone (e.g., definitions of abbreviations, copyright attribution). Not all figures include notes. Notes are flush left, non-italicised. If present they begin with Note. (italicised, period ending). The notes area will include reference information if not an original figure, and copyright information as required.
General rules
- In the text, refer to every figure by its number, no italics, but with a capital "F" for "Figure". For example, "As shown in Figure 1, ..."
- There are two options for the placement of figures in a paper. The first option is to place all figures on separate pages after the reference list. The second option is to embed each figure within the text.
- If you reproduce or adapt a figure from another source (e.g., an image you found on the internet), you should include a copyright attribution in the figure note, indicating the origin of the reproduced or adapted material, in addition to a reference list entry for the work. Include a permission statement (Reprinted or Adapted with permission) only if you have sought and obtained permission to reproduce or adapt material in your figure. A permission statement is not required for material in the public domain or openly licensed material. For student course work, AUT assignments and internal assessments, a permission statement is also not needed, but copyright attribution is still required.
- Important note for postgraduate students and researchers: If you wish to reproduce or adapt figures that you did not create yourself in your thesis, dissertation, exegesis, or other published work, you must obtain permission from the copyright holder/s, unless the figure is in the public domain (copyright free), or licensed for use with a Creative Commons or other open license. Works under a Creative Commons licence should be cited accordingly. See Using works created by others for more information.
Please check the APA style website for an illustration of the basic figure component & placement of figure in a text.
More information & examples from the APA Style Manual , s. 7.22-7.36, pp. 225–250
Figure reproduced in your text
Note format - for notes below the figure

In-text citation:
Reference list entry:
Referring to a figure in a book
If you refer to a figure included in a book but do not include it in your text, format the in-text citation and the reference list entry in the usual way, citing the page number where the figure appears.
Note format - for notes below the figure

Referring to a figure in an article
If you refer to a figure in an article but do not include it in your text, format the in-text citation and the reference list entry in the usual way for an article, citing the page number where the figure appears.
Note format - for notes below the figure

Reference list:

Referring to a figure on a webpage
If you refer to a figure on a webpage and do not include it in your text, format the in-text citation and the reference list entry in the usual way for a webpage,
Not every reference to an artwork needs a reference list entry. For example, if you refer to a famous painting, as below, it would not need a reference.
Finding image details for your figure caption or reference
- clicking on or hovering your mouse over the image
- looking at the bottom of the image
- looking at the URL
- If there is no title, create a short descriptive one yourself and put it in square brackets e.g. [...]
- For more guidance, see Visual works
If it has been formally published reference your work as you would any other published work.
If the work is available on a website reference it as a webpage (see examples in the webpage section ).
Citing your own figures, graphs or images in an assignment:
- Include the title
- Add a note explaining the content. No copyright attribution is required.
- You can, if you wish, add a statement that it is your own work
- You do not need an in-text citation or add it to your reference list
- See example in APA manual p.247, Figure 7.17 Sample photograph
Great Barrier Island

Note. Photo of Great Barrier Island taken from Orewa at sunrise. Own work.
- << Previous: Reports & grey literature
- Next: Tables >>
- Last Updated: Mar 5, 2024 3:25 PM
- URL: https://aut.ac.nz.libguides.com/APA7th

MLA Citation Guide (9th Edition): Images, Charts, Graphs, & Tables
- Advertisements
- Audio Materials
- Books, Ebooks, & Book Chapters
- Class Materials (Notes, Slides, & Recordings)
- Creative Commons Works
- Encyclopedias, & Dictionaries
- Images, Charts, Graphs, & Tables
- Journal Articles
- Magazine Articles
- Newspaper Articles
- Personal Communications (Interviews, Emails, etc.)
- Religious Works
- Social Media
- Websites (includes documents/PDFs posted online)
- When Information Is Missing
- When a Work Is Quoted in Another Source
- Permalinks, URLs, & DOIs!
- Quoting vs. Paraphrasing
- Works Cited & Paper Format
- Citation Tools
- Citation Managers
Reproducing vs. Just Citing
This happens if you only cite information from an image, infographic, chart, table, or graph and do not reproduce it in your paper. If you're only citing information from an image, infographic, Chart, Table or Graph:
- Provide an in-text citation. Use the citation format of the source where the image is found. (e.g., if you find the image on a website, use the in-text citation of a website).
- Cite the image in your Works Cited List. Use the citation format of the source where the image is found. (e.g., if you found the image on a website, cite the website).
Reproducing happens when you copy or recreate an image, infographic, table, graph, or chart that is not your original creation. If you reproduce one of these works in your assignment, you must create a note (or "caption") underneath the photo, image, chart, graph, or table to show where you found it. If you do not refer to it anywhere else in your assignment, you do not have to include the citation for this source in a Works Cited list.
Inserting a Table You Reproduced
- Start by adding a label for your table (e.g., Table 1, bolded and aligned to the left) followed by a description of what information is contained in the table.
- Below the table, add the word Adapted from: followed by the full citation for the source where you found the information. For example, if you found the information on a website, use the Works Cited list citation format for citing a website. For sources with individual authors, do not invert the first and last names at the beginning of the citation.
- If the table is not cited in the text of your assignment, you do not need to include it in your Works Cited list.
Variables in determining victims and aggressors
Adapted from: Andrea Mohr. "Family Variables Associated With Peer Victimization." Swiss Journal of Psychology, vol . 65, no. 2, 2006, pp. 111. Gale Psychology Collection , https://doi.org/10.1024/1421-0185.65.2.107 . PDF download.
Your Photographs & Images
If you reproduce your own photograph or image in your coursework, you do not need to cite it. However, Simmons Library recommends adding a figure note beneath the image that reads "Photograph by author" or "Image by author."
Inserting a Table You Adapted from Multiple Sources
- Start by adding a label for your table (e.g., Table 1, bolded) followed by a description of what information is contained in the table.
- Below the table, add the word Adapted from: followed by the full citation for the sources where you found the information. For example, if you found the information on a website, use the Works Cited list citation format for citing a website. For sources with individual authors, do not invert the first and last names at the beginning of the citation.
- List your sources in alphabetical order by the author's last name. Separate each source with a semi-colon (;).
Total downloads (in millions) of communication apps Discord, Telegram and WeChat through Apple App store and Google Play store in September 2020
Adapted from: Airnow. "Leading communication apps in the Google Play Store worldwide in September 2020, by number of downloads." Statista , Oct. 2020.; Airnow. "Leading social networking apps in the Apple App Store worldwide in September 2020, by number of downloads." Statista , Oct. 2020.
Inserting an Image Reproduced from a Source
If you are recreating visual material which is not a table (e.g., infographic, maps, photo, graph):
- Under the image, add a figure number (e.g., Fig. 1.) and short description.
- Add the full citation after the description. Follow the citation template for your source. For example, if you're citing an infographic from a website, use the template for citing infographics posted on a website. For sources with individual authors, do not invert the first and last names at the beginning of the citation.
- If the image is not cited in the text of your assignment, you do not need to include it in your Works Cited list.
Fig. 1. Annie Green. "Yoga: Stretching Out." Sports Digest, 8 May 2006, p. 22.

Fig. 2. Pauline Cheung. "Short Skirt S/S/ 15 China Womenswear Commercial Update." WGSN , 4 June 2016, p. 2.
- << Previous: Encyclopedias, & Dictionaries
- Next: Journal Articles >>
- Last Updated: Feb 28, 2024 1:45 PM
- URL: https://simmons.libguides.com/mla
APA Citation Style, 7th edition: Figures
- General Style Guidelines
- One Author or Editor
- Two Authors or Editors
- Three to Five Authors or Editors
- Article or Chapter in an Edited Book
- Article in a Reference Book
- Edition other than the First
- Translation
- Government Publication
- Journal Article with 1 Author
- Journal Article with 2 Authors
- Journal Article with 3–20 Authors
- Journal Article 21 or more Authors
- Magazine Article
- Newspaper Article
- Basic Web Page
- Web page from a University site
- Web Page with No Author
- Entry in a Reference Work
- Government Document
- Film and Television
- Youtube Video
- Audio Podcast
- Electronic Image
- Twitter/Instagram
- Lecture/PPT
- Conferences
- Secondary Sources
- Citation Support
- Avoiding Plagiarism
- Formatting Your Paper
About Citing Sources
For each type of source in this guide, both the general form and an example will be provided.
The following format will be used:
In-Text Citation (Paraphrase) - entry that appears in the body of your paper when you express the ideas of a researcher or author using your own words. For more tips on paraphrasing check out The OWL at Purdue .
In-Text Citation (Quotation) - entry that appears in the body of your paper after a direct quote.
References - entry that appears at the end of your paper.
When you use a figure in your paper that has been adapted or copied directly from another source, you need to reference the original source. This reference appears as a caption underneath the figure that you copied or adapted for your paper.
Any image that is reproduced from another source also needs to come with copyright permission; it is not enough just to cite the source.
- Number figures consecutively throughout your paper.
- Figures should be labeled "Figure (number)" ABOVE the figure.
- Double-space the caption that appears under a figure.
General Format 1 (Figure from a Book):
Subject Guide

- << Previous: Electronic Image
- Next: Social Media >>

- Last Updated: Feb 6, 2024 11:45 AM
- URL: https://guides.himmelfarb.gwu.edu/APA

- Himmelfarb Intranet
- Privacy Notice
- Terms of Use
- GW is committed to digital accessibility. If you experience a barrier that affects your ability to access content on this page, let us know via the Accessibility Feedback Form .
- Himmelfarb Health Sciences Library
- 2300 Eye St., NW, Washington, DC 20037
- Phone: (202) 994-2850
- [email protected]
- https://himmelfarb.gwu.edu
Thank you for visiting nature.com. You are using a browser version with limited support for CSS. To obtain the best experience, we recommend you use a more up to date browser (or turn off compatibility mode in Internet Explorer). In the meantime, to ensure continued support, we are displaying the site without styles and JavaScript.
- View all journals
- My Account Login
- Explore content
- About the journal
- Publish with us
- Sign up for alerts
- Data Descriptor
- Open access
- Published: 26 June 2020
Building a PubMed knowledge graph
- Jian Xu ORCID: orcid.org/0000-0003-4886-4708 1 ,
- Sunkyu Kim 2 ,
- Min Song 3 ,
- Minbyul Jeong 2 ,
- Donghyeon Kim 2 ,
- Jaewoo Kang ORCID: orcid.org/0000-0001-6798-9106 2 ,
- Justin F. Rousseau ORCID: orcid.org/0000-0002-2817-9124 4 ,
- Xin Li ORCID: orcid.org/0000-0002-8169-6059 5 ,
- Weijia Xu 6 ,
- Vetle I. Torvik 7 ,
- Chongyan Chen 5 ,
- Islam Akef Ebeid 5 ,
- Daifeng Li 1 &
- Ying Ding ORCID: orcid.org/0000-0003-2567-2009 4 , 5
Scientific Data volume 7 , Article number: 205 ( 2020 ) Cite this article
34k Accesses
81 Citations
37 Altmetric
Metrics details
- Communication and replication
- Data integration
- Data mining
PubMed ® is an essential resource for the medical domain, but useful concepts are either difficult to extract or are ambiguous, which has significantly hindered knowledge discovery. To address this issue, we constructed a PubMed knowledge graph (PKG) by extracting bio-entities from 29 million PubMed abstracts, disambiguating author names, integrating funding data through the National Institutes of Health (NIH) ExPORTER, collecting affiliation history and educational background of authors from ORCID ® , and identifying fine-grained affiliation data from MapAffil. Through the integration of these credible multi-source data, we could create connections among the bio-entities, authors, articles, affiliations, and funding. Data validation revealed that the BioBERT deep learning method of bio-entity extraction significantly outperformed the state-of-the-art models based on the F1 score (by 0.51%), with the author name disambiguation (AND) achieving an F1 score of 98.09%. PKG can trigger broader innovations, not only enabling us to measure scholarly impact, knowledge usage, and knowledge transfer, but also assisting us in profiling authors and organizations based on their connections with bio-entities.
Machine-accessible metadata file describing the reported data: https://doi.org/10.6084/m9.figshare.12452597
Similar content being viewed by others

Assessing GPT-4 for cell type annotation in single-cell RNA-seq analysis
Wenpin Hou & Zhicheng Ji
A visual-language foundation model for computational pathology
Ming Y. Lu, Bowen Chen, … Faisal Mahmood

scGPT: toward building a foundation model for single-cell multi-omics using generative AI
Haotian Cui, Chloe Wang, … Bo Wang
Background and Summary
Experts in healthcare and medicine communicate in their own languages, such as SNOMED CT, ICD-10, PubChem, and gene ontology. These languages equate to gibberish for laypeople, but for medical minds, they are an intricate method of transporting important semantics and consensus capable of translating diagnoses, medical procedures, and medications among millions of physicians, nurses, and medical researchers, thousands of hospitals, hundreds of pharmacies, and a multitude of health insurance companies. These languages (e.g., genes, drugs, proteins, species, and mutations) are the backbone of quality healthcare. However, they are deeply embedded in publications, making literature searches increasingly onerous because conventional text mining tools and algorithms continue to be ineffective. Given that medical domains are deeply divided, locating collaborators across domains is arduous. For instance, if a researcher wants to study ACE2 gene related to COVID-19, he or she would like to know the following: which researchers are currently actively studying ACE2 gene, what are the related genes, diseases, or drugs discussed in these articles related to ACE2 gene, and with whom could the researcher collaborate? This is a strenuous position to be in, and the aforementioned problems diminish the curiosity directed at the topic.
Many studies have been devoted to building open-access datasets to solve bio-entity recognition problems. For example, Hakala et al . 1 used a conditional random field classifier-based tool to recognize the named entities from PubMed and PubMed Central. Bell et al . 2 performed a large-scale integration of a diverse set of bio-entities and their relationships from both bio-entity datasets and PubMed literature. Although these open-access datasets are predominantly about bio-entity recognition, researchers have also been interested in extracting other types of entities and relationships from PubMed, including the mapping of author affiliations to cities and their geocodes 3 , 4 , author name disambiguation 5 (AND), and author background information collections 6 . Although the focus of previous research has been on limited types of entities, the goal of our study was to integrate a comprehensive dataset by capturing bio-entities, disambiguated authors, funding, and fine-grained affiliation information from PubMed literature.
Figure 1 illustrates the bio-entity integration framework. This framework consists of two parts: (1) bio-entity extraction, which contains entity extraction, named entity recognition (NER), and multi-type normalization, and (2) integration, which connects authors, ORCID, and funding information.
Bio-entity integration framework for PKG.
The process illustrated in Fig. 1 can be described as follows. First, we applied the high-performance deep learning method Bidirectional Encoder Representations from Transformers for Biomedical Text Mining (BioBERT) 7 , 8 to extract bio-entities from 29 million PubMed abstracts. Based on the evaluation, this method significantly outperformed the state-of-the-art methods based on the F1 score (by 0.51%, on average). Then, we integrated two existing high-quality author disambiguation datasets: Author-ity 5 and Semantic Scholar 9 . We obtained the disambiguated authors of PubMed articles with full coverage and quality of 98.09% in terms of the F1 score. Next, we integrated additional fields from credible sources into our dataset, which included the projects funded by the National Institutes of Health (NIH) 10 , the affiliation history and educational background of authors from ORCID 6 , and fine-grained region and location information from the MapAffil 2016 dataset 11 . We named this new interlinked dataset “PubMed Knowledge Graph” (PKG). PKG is by far the most comprehensive, up-to-date, high-quality dataset for PubMed regarding bio-entities, articles, scholars, affiliations, and funding information. Being an open dataset, PKG contains rich information ready to be deployed, facilitating the effortless development of applications such as finding experts, searching bio-entities, analyzing scholarly impacts, and profiling scientists’ careers.
Bio-entity extraction
The bio-entity extraction component has two models: (1) an NER model, which recognizes the named entities in PubMed abstracts based on the BioBERT model 7 , and (2) a multi-type normalization model, which assigns unique IDs to recognize biomedical entities.
Named Entity Recognition (NER)
The NER task recognizes a variety of domain-specific proper nouns in a biomedical corpus and is perceived as one of the most notable biomedical text mining tasks. In contrast to previous studies that have built models based on long short-term memory (LSTM) and conditional random fields (CRFs) 12 , 13 , the recently proposed Bidirectional Encoder Representations from Transformers (BERT) 14 model achieves excellent performance for most of the NLP tasks with minimal task-specific architecture modifications. The transformers applied in BERT connect the encoders and decoders through self-attention for greater parallelization and reduced training time. BERT was designed as a general-purpose language representation model that was pre-trained on English Wikipedia and BooksCorpus. Consequently, it is incredibly challenging to maintain high performance when applying BERT to biomedical domain texts that contain a considerable number of domain-specific proper nouns and terms (e.g., BRCA1 gene and Triton X-100 chemical). BERT required refinement, so BioBERT—a neural network-based high-performance NER model—was developed. Its purpose is to recognize the known biomedical entities and discover new biomedical entities.
First, in the NER component, the case-sensitive version of BERT is used to initialize BioBERT. Second, PubMed articles and PubMed Central articles are used to pre-train BioBERT’s weights. The pre-trained weights are then fine-tuned for the NER task. While fine-tuning BERT (BioBERT), we used WordPiece tokenization 15 to mitigate the out-of-vocabulary issue. WordPiece embedding is a method of dividing a word into several units (e.g., Immunoglobulin divided into I ##mm##uno ##g ##lo ##bul ##in) and expressing each unit. This technique is effective at extracting the features associated with uncommon words. The NER models available in BioBERT can predict the following seven tags: IOB2 tags (i.e., Inside, Outside, and Begin) 16 , X (i.e., a sub-token of WordPiece), [CLS] (i.e., the leading token of a sequence for classification), [SEP] (i.e., a sentence delimiter), and PAD (i.e., a padding of each word in a sentence). The NER models were fine-tuned as follows 8 :
where k represents the indexes of seven tags {B, I, O, X, [CLS], [SEP], PAD}, p is the probability distribution of assigning each k to token i , and \({T}_{i}\in {R}^{H}\) is the final hidden representation, which is calculated by BioBERT for each token i . H is the hidden size of T i , \(W\in {R}^{K\times H}\) is a weight matrix between k and T i , K represents the number of tags and is equal to 7, and b is a K-dimensional vector that records the bias on each k . The classification loss L is calculated as follows:
where Θ represents the trainable parameters, and N is the sequence length.
First, a tokenizer was applied to words in a sentence on a dataset with labels in the CoNLL format 17 . The WordPiece algorithm was then applied to the sub-words of each word. Consequently, BioBERT was able to extract diverse types of bio-entities. Furthermore, an entity or two entities with frequently-occurring token interaction would be marked with more than one entity type span (26.2% for all PubMed abstracts). Based on the calculated probability distribution, we were able to choose the correct entity type when entities were tagged with more than two types according to the probability-based decision rules 8 .
Multi-type normalization
Because an entity may be referred to by several synonymous terms (synonyms), and a term can be polysemous if it refers to multiple entity types (polysemy), we require a normalization process for the extracted entities. However, it is a daunting challenge to build a single normalization tool for multiple entity types because there exist various normalization models that depend on the type of entity. We addressed this issue by combining multiple NER normalization models into one multi-type normalization model that assigns IDs to extracted entities. Table 1 illustrates the statistics of the proposed normalization model.
The multi-type normalization model is based on a normalization model per entity type (Table 1 ). To improve the number of normalized entities, we added the disease names from the PolySearch2 dictionary (76,001 names of 27,658 diseases) to the sieve-based entity linking dictionary (76,237 names of 11,915 diseases). We also added the drug names from DrugBank 18 and the U.S. Food and Drug Administration (FDA) to the tmChem dictionary. Because there are no existing normalization models for species, we normalized species based on dictionary lookup. Using tmVar 2.0, we created a dictionary of mutations with normalized mutation names, in which a mutation with several names was assigned to one normalized name or ID.
Author Name Disambiguation (AND)
Despite a rigorous effort to create global author IDs (e.g., ORCID and ResearcherID), most articles in PubMed, particularly those before 2003 (the year in which the field ORCID was added into PubMed), provide limited author information with respect to last name, first initial, and affiliation (only for first authors before 2014). Author information is not effective meta-data to be used directly as a unique identifier because different people may have the same names, and the names and affiliations of an individual can change over time. AND is essential for identifying unique authors.
In recent decades, researchers have made several attempts to solve the AND problem, using three types of methods. The first type of method relies on manual matching of articles with authors by surveying scientists or consulting curricula vitae (CVs) gathered from the Internet 19 . Although this type of method ensures high accuracy, a considerable amount of investment in labor is required to collect and code the data, which is impractical for huge datasets. The second type of method uses publicly-accessible registry platforms, such as ORCID or Google Scholar, to help researchers identify their own publications, which produces a source of highly accurate and low-cost accessible disambiguation of authorship for large numbers of authors. However, registries cover only a small proportion of researchers 20 , 21 , which introduces a form of survivor bias into samples. The third type of method uses an automated approach to estimate the similarity of author instance feature combinations and identify whether they refer to the same person. The features for automated AND include author name, author affiliation, article keywords, journal names 22 , coauthor information 23 , and citation patterns 24 . Automated methods typically rely on supervised or unsupervised machine learning, in which the machine learns how to weigh the various features associated with author names and where to assign a pair of author names either to the same author or to two different authors 25 , 26 . This type of method can potentially avoid the shortcomings of the previous two types. Moreover, automated methods have been improved to a high level of accuracy after years of development.
For PubMed, automated methods are the optimal choice because they can overcome the shortcomings of the other two methods while simultaneously providing high-quality AND results for the entire dataset. Several scholars have disambiguated the authors using automated methods. Although the evaluations of these results have exhibited different levels of accuracy and coverage limitations, we believe that integrating them with due diligence can yield a high-quality AND dataset with full coverage of PubMed articles.
According to our investigation, a high-quality PubMed AND dataset with complete coverage can be obtained through the integration of the following two existing AND datasets:
Author-ity: The Author-ity database uses diverse information about authors and publications to determine whether two or more instances of the same name (or of highly similar names) on different papers represent the same person. According to the AND evaluation based on the method discussed in the section Technical Validation , the F1 score of Author-ity is 98.16%, which is the highest accuracy result that we have observed. However, this dataset only covers authors before 2009.
Semantic Scholar: The Semantic Scholar database trains a binary classifier to merge a pair of author names and use the pair to create author clusters incrementally. According to the AND evaluation based on the method discussed in the section Technical Validation , the F1 score of Semantic Scholar is 96.94%, which is 1.22% lower than that of Author-ity. However, it has the most comprehensive coverage of authors.
Because the Author-ity dataset has a higher F1 score than the Semantic Scholar dataset, we selected the author’s unique ID of the Author-ity dataset as the primary AND_ID. AND_ID is limited by time range (containing PubMed papers before 2009); however, we supplemented authors after 2009 using the AND result from Semantic Scholar. The following steps were applied:
Step 1: We allocated the author’s unique ID to each author instance according to the Author-ity AND results such that authors from the Author-ity dataset (before 2009) have unique author IDs.
Step 2: For authors that have the same Semantic Scholar AND_ID but never appear in the Author-ity dataset, we generated a new AND_ID to label them. For example, author “Pietranico R.” published two papers in 2012 and 2013 and had two corresponding author instances. Because all papers that “Pietranico R.” published were after 2009, they were not covered by Author-ity and therefore had no AND_ID allocated by Author-ity. However, the authors disambiguated correctly by Semantic Scholar were allocated unique AND_IDs in Semantic Scholar. To maintain the consistency in labeling, we generated a new AND_ID continuing AND_IDs of Author-ity to label these two author instances as disambiguated by Semantic Scholar.
Step 3: For author instances with a unique AND_ID in Semantic Scholar and in which authors (at least one) had the same Author-ity AND_ID, we allocated the Author-ity AND_ID to all author instances as their unique ID. For example, “Maneksha S.” published three papers in 2007, 2009, and 2010, and the first two author instances had a unique Author-ity AND_ID. However, the last one had no Author-ity AND_ID because it was beyond the time coverage of the Author-ity dataset. Nevertheless, based on the AND results of Semantic Scholar, the three author instances had an identical AND_ID. Therefore, the last author instance with no Author-ity AND_ID could be labeled with the same ID as the other two author instances.
Extended multi-source information integration
In addition to bio-entity extraction by BioBERT and AND, we made a considerable effort to integrate PubMed by extending multi-source data into PKG, which exploited the mapping connections between AND_ID and the PubMed identifier (PMID) to build relationships between different objects to provide a comprehensive overview of the PubMed dataset. These integrated data include the funding data from NIH ExPORTER, the affiliation history and educational background of authors from ORCID, and the fine-grained region and location information from the MapAffil 2016 dataset. The entities and their associated relationships are depicted in Fig. 2 .
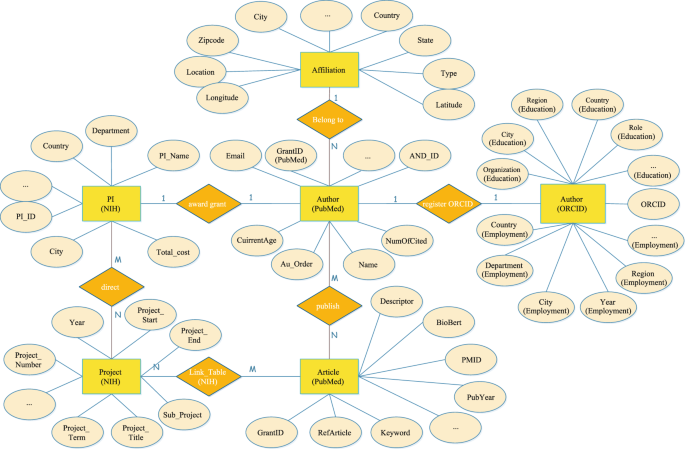
Entities and relationships in PKG.
Project data from NIH ExPORTER
NIH ExPORTER provides data files that contain research projects funded by major funding agencies such as the Centers for Disease Control and Prevention (CDC), the NIH, the Agency for Healthcare Research and Quality (AHRQ), the Health Resources and Services Administration (HRSA), the Substance Abuse and Mental Health Services Administration (SAMHSA), and the U.S. Department of Veterans Affairs (VA). Furthermore, it provides publications and patents citing support from these projects. It consists of 49 data fields, including the amount of funding for each fiscal year, organization information of the PIs, and the details of the projects. According to our investigation, NIH-funded research accounts for 80.7% of all grants recorded in PubMed.
The NIH ExPORTER dataset contains a unique PI_ID for each scholar who received NIH funding between 1985 and 2018, and his or her PMIDs of the published articles. Through the mapping of PMIDs in NIH ExPORTER to PMIDs in PubMed, 1:N connections between the PI and articles have been established, paving the way for investigating the article details of a specific PI, and vice versa. Furthermore, by mapping PI names (last name, first initial, and affiliation) to author names that were listed in articles supported by the PI’s projects, a 1:1 connection between the PI and the AND_ID was established, providing a way to obtain PI-related article information, regardless of whether the article was labeled with a project ID.
Employment history and educational background data from ORCID
According to its website, “ORCID is a nonprofit organization helping to create a world in which all who participate in research, scholarship, and innovation are uniquely identified and connected to their contributions and affiliations across disciplines, borders, and time” 27 . It maintains a registry platform for researchers to actively participate in identifying their own publications, information about formal employment relationships with organizations, and educational backgrounds. ORCID provides an open-access dataset called ORCID Public Dataset 2018 6 , which contains a snapshot of all public data in the ORCID Registry associated with an ORCID record that was created or claimed by an individual as of October 1, 2018. The dataset includes 7,132,113 ORCID iDs, of which 1,963,375 have educational affiliations and 1,913,610 have employment affiliations.
As a result of the proliferation of ORCID identifiers, PubMed has used ORCID identifiers as alternative author identifiers since 2013 28 . Using the following two steps, we could map ORCID records to the PubMed authors. Our first step was to map the author instances in PubMed to an ORCID record based on the feature combinations of article DOI and author name (last name and first initial). Because the DOI is not a compulsory field for PubMed, we appended the feature combinations of article titles, journals, and author names to map the records between the two datasets. The result contained many 1:1 connections between a disambiguated author of PubMed and an ORCID record. Furthermore, 1:1 connections between AND_ID and ORCID iD, and 1:N connections between AND_ID and background information (education and employment) were established.
Fine-grained affiliation data
The MapAffil 2016 dataset 3 resolves PubMed authors’ affiliation strings to cities and associated geocodes worldwide. This dataset was constructed based on a snapshot of PubMed (which included the Medline and PubMed-not-Medline records) acquired in the first week of October 2016. Affiliations were linked to a specific author on a specific article. Prior to 2014, PubMed only recorded the affiliation of the first author. However, MapAffil 2016 covered some PubMed records that lacked affiliations and were harvested elsewhere, such as from PMC, NIH grants, the Microsoft Academic Graph, and the Astrophysics Data System. All affiliation strings were processed using MapAffil to identify and disambiguate the most specific place names. The dataset provides the following fields: PMID, author order, last name, first name, year of publication, affiliation type, city, state, country, journal, latitude, longitude, and Federal Information Processing Standards (FIPs) code.
The MapAffil 2016 dataset does have a limitation because it does not cover the PubMed data after 2015 (covering 62.9% affiliation instances in PubMed). Consequently, we performed an additional step to improve the fraction of coverage. We collected authors (who published their first article before 2016 and continued publishing articles after 2015) by their AND_IDs. The new affiliation instances of the author after 2015 succeeded their corresponding fine-grained affiliation data from the affiliation instances before 2016 (fraction of affiliation instance coverage increased to 84.2%) if the author did not change affiliation. We also applied an up-to-date open-source library Affiliation Parser 4 to extract additional fine-grained affiliation fields from all affiliation instances, including department, institution, email, ZIP code, location, and country.
Table 2 summarizes the date coverage and version information of integrated datasets and open-access software used to extract data.
Data Records
We built PKG with bio-entities extracted from PubMed abstracts, AND results of PubMed authors, and the integrated multi-source information. This dataset is freely available on Figshare 29 . It contains seven comma-separated value (CSV) files named “Author_List,” “Bio_entities_Main,” “Bio_entities_Mutation,” “Affiliations,” “Researcher_Employment,” “Researcher_Education,” and “NIH_Projects”. The details are presented in Table 3 . PubMed raw data are not included into Figshare file set because the amount of PubMed raw data is too large and they are not generated or altered by our methods. PubMed raw data can be freely downloaded from PubMed website 30 . We also provide the following download link ( http://er.tacc.utexas.edu/datasets/ped ), which contains both the PubMed raw data and PKG dataset to facilitate the application of PKG dataset.
The statistics of all five types of extracted entities are presented in Table 4 .
Each data field is self-explanatory by its name, and fields with the same name in other tables follow the same data format that can be linked across tables. Tables 5 – 11 illustrate the field name, format, and short description of fields for each data file listed in Table 3 .
Updating PKG is a complex task because it is subject to the update of different data sources and requires significant computation. In the future, we hope to refresh PKG quarterly based on PubMed updated files and updated datasets from other sources. We may also develop an integrative ontology to integrate all types of entities.
Technical Validation
Validity of bio-entity extraction.
To validate the performance of the bio-entity extraction, we established BERT and the state-of-the-art models as baselines. Then, we calculated the entity-level precision, recall, and F1 scores of these models as evaluation metrics. The datasets and the test results of biomedical NER are presented in Table 12 .
In Table 12 , we report the precision (P), recall (R), and F1 (F) scores of each dataset. The highest scores are in boldface , and the second-highest scores are underlined . Sachan et al . 31 reported the scores of the state-of-the-art models for the NCBI disease and BC2GM datasets, presented in Table 10 . Moreover, the scores for the 2010 i2b2/VA dataset were obtained from Zhu et al . 32 (single model), and the scores for the BC5CDR and JNLPBA datasets were obtained from Yoon et al . 13 . The scores for the BC4CHEMD dataset were obtained from Wang et al . 33 , and scores for the LINNAEUS and Species-800 datasets were obtained from Giorgi and Bader 34 .
According to Table 12 , BERT, which is pre-trained on the general domain corpus, was highly effective. On average, the state-of-the-art models outperformed BERT by 2.28% in terms of the F1 score. However, BioBERT obtained the highest F1 score in recognizing Genes/Proteins, Diseases, and Drugs/Chemicals. It outperformed the state-of-the-art models by 0.51% in terms of the F1 score, on average.
Validity of multi-type entity normalization
We used the multi-type normalization model to assign unique IDs to synonymous entities. Table 13 presents the performance of the multi-type entity normalization model.
As shown in Table 13 , with respect to genes and proteins, there were 75 different species in the BC3 Gene Normalization (BC3GN) test set, but GNormPlus focused only on seven of these species. Consequently, GNormPlus achieved a considerably lower F1 score by 36.6% on the multispecies test set (BC3GN) than on the human species test set (BC2GN). For mutations, tmVar 2.0 achieved F1 scores close to 90% on two corpora: OSIRISv1.2 and the Thomas corpus.
Validity of author name disambiguation
The validation of author disambiguation remains a challenge because there is a lack of abundant validation sets. We applied a method using the NIH ExPORTER-provided information on NIH-funded researchers to evaluate the precision, recall, and F1 measures of the author disambiguation 35 .
NIH ExPORTER provides information about the principal investigator ID (PI_ID) for each scholar who received NIH funding between 1985 and 2018. Because applicants established a unique PI_ID and used the PI_ID across all grant applications, these PI_IDs have extremely high fidelity. NIH ExPORTER also provides article PMIDs as project outputs, which can be conveniently used as a connection between PI_IDs and AND_ID.
We confirmed the bibliographic information of the NIH-funded scientists who received NIH funding during the years 1985–2018. Our AND evaluation steps were as follows: First, we collected project data for the years 1981–2018 in NIH ExPORTER, including 304,782 PI_ID records and the corresponding 331,483 projects. Next, we matched the projects to articles acknowledging support by the grant, which were also recorded in the NIH ExPORTER dataset. We matched 214,956 of the projects to at least one article and identified 1,790,949 articles funded by these projects. Some of these projects (116,527) did not match articles and were excluded. Because the NIH occasionally awards a project to a team that includes more than one PI, we eliminated the 13,154 records that contained multiple PIs because they could result in uncertain credit allocation. Consequently, our relevant set of PIs decreased to 147,027 individuals associated with 1,749,873 articles and 201,802 projects.
We then connected NIH PI_IDs from NIH ExPORTER to AND_IDs using the article PMIDs and author (PI)’s last name plus the initials as a crosswalk. This step resulted in 1,400,789 unique articles remaining, associated with 109,601 PI_IDs and 107,380 AND_IDs. Finally, we computed precision (P) based on the number of articles associated with the most frequent AND_ID-to-PI_ID matched over the number of all articles associated with a specific AND_ID 36 . Furthermore, we computed recall (R) based on the number of articles associated with the most frequent PI_ID-to-AND_ID matched over the number of all articles associated with a particular PI_ID 36 . Figure 3 summarizes the precision, recall, and F1 calculations.
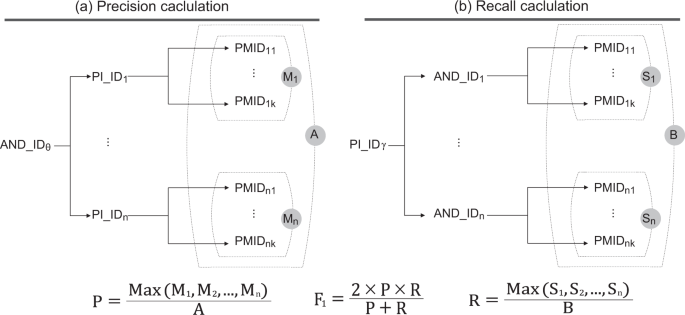
Calculation of Precision, Recall, and F1 Score.
Table 14 illustrates the precision, recall, and F1 scores for Author-ity, Semantic Scholar, and our integrated AND result.
As presented in Table 14 , after integrating the AND results of Author-ity and Semantic Scholar, we obtained a high-quality integrated AND result that outperformed Semantic Scholar by 1.15% in terms of the F1 score and had more comprehensive coverage (until 2018) than Author-ity (until 2009).
The evaluation results of AND might be slightly overestimated. The PIs of NIH grants usually have many publications over a long period and might be more likely to have rich information, such as affiliations and email addresses, about publications. Therefore, it should be easier to acquire higher performance on AND tasks than that of new entrants who published fewer papers and may lack of sufficient information for AND. Furthermore, approximately 1.15% of the author instances cannot be disambiguated since they do not exist in the Author-ity or Semantic Scholar AND results, which further slightly reduces the performance of AND results theoretically. However, the Semantic Scholar AND results and the AND Integration are evaluated based on the same baseline dataset with Author-ity in this section, and the evaluation of Author-ity performance using a random sample of articles indicates reliably high quality: the recall of the Author-ity dataset is 98.8%, the lumping (putting two different individuals into the same cluster) of the Author-ity dataset affects 0.5% of the clusters, and the splitting (assigning articles written by the same individual to more than one cluster) of the Author-ity dataset affects 2% of the articles 5 . Consequently, we believe these factors have a limited impact on AND performance.
Usage Notes
Networking and collaboration have been associated with faculty promotions in academic medical centers 37 . Barriers exist for identifying researchers working on common bio-entities to facilitate collaboration. It is a challenge even at a single academic institution to identify potential collaborators who are working on the same bio-entities. This has led to many institution-specific projects profiling the faculty associated with the topics that they are studying 38 , 39 , 40 , 41 . The challenge is exacerbated when we search across multiple institutions.
Researchers, academic institutions, and the pharmaceutical industry often face the challenge of identifying researchers working on a specific bio-entity. A traditional bibliographic database specializes only in returning an enormous number of related articles for particular keyword or term searches. Bio-entity profiling for researchers offers an advantage over this traditional approach by identifying specific connections between bio-entities and disambiguated authors, in which bio-entity profiling for researchers can directly locate the core specialists whose research is focused on these bio-entities. Furthermore, a bipartite author-entity network projection analysis can identify a specific author’s neighborhood with similar research interest, which is crucial for community detection and collaborative recommendations.
We sought to use the PKG dataset to understand the trends over time of researcher-centric and bio-entity-centric activity by the following use cases: (1) researcher-centric for Stephen Silberstein, MD, a neurologist and expert in headache research; (2) calcitonin Gene-Related Peptide (CGRP), a target of inhibition for one of the newest therapeutics in migraine treatment; and (3) bipartite author-entity projection network analysis for coronavirus, a disease that causes respiratory illness with symptoms such as a fever, cough, and difficulty breathing.
For researcher-centric and bio-entity-centric activities, we collected 455 articles with Dr. Silberstein as an author and 7,877 articles on CGRP in the PKG dataset from 1970 to 2018 and extracted the bio-entities from these articles. Several publications and bio-entities were used for profiling the career of Dr. Silberstein. Several publications and the author’s distribution were used for profiling CGRP. For bipartite author-entity projection network analysis, we collected 9,778 articles on coronavirus in the PKG dataset from 1969 to 2019.
Researcher-centric activity
For Dr. Silberstein, 539 bio-entities, including 342 diseases, 142 drugs, 24 genes, 17 species, and 14 mutations, were extracted from 455 articles. As depicted in Fig. 4(a) , “Headache” and “migraine” were his two most studied diseases, reaching 21 and 19 articles, respectively, in 2004. We trended his research over time on triptans, starting with sumatriptan. CGRP began to emerge in his publications starting in 2015. We noted the five researchers that have collaborated with Dr. Silberstein through his career and map with PKG their collaborations, interactions, and institutions over time. Visualizing the profiles of individual researchers can help to understand the trends in their topics of interest and collaboration patterns to enable an understanding of collaboration factors that may be associated with academic success or scientific discovery.
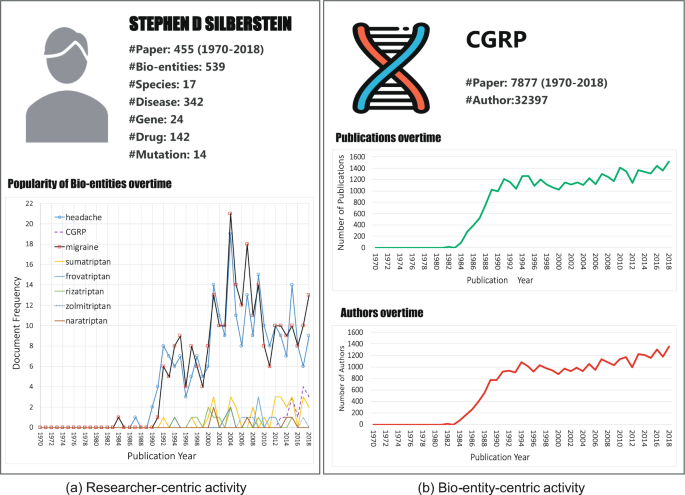
Trends over time of researcher-centric and bio-entity-centric activity.
Bio-entity-centric activity
For CGRP, there are currently 7,877 articles by 32,392 authors on CGRP dating back to 1982. Figure 4(b) illustrates that there was a dramatic increase in the number of CGRP-related articles, from 13 in 1982 to 1,209 in 1991, with a steady increase to 1,517 in 2018. The trend of the number of authors over time was similar to that of the volume of articles on CGRP.
As we demonstrated with a previous analysis of the repurposing of Aspirin 42 , 43 , we observe research on CGRP starting at approximately the same time as the research on triptans for the treatment of migraines. Research on the pathophysiology of migraines identified a central role of the neuropeptide calcitonin gene-related peptide (CGRP), which is thought to be involved with the dilation of cerebral and dural blood vessels, release of inflammatory mediators, and the transmission of pain signals 44 . Research on the mechanism of the action of triptans—serotonin receptor agonists—has led to an understanding that they normalize elevated CGRP levels, which among other mechanisms, has led to an improvement in migraine headache symptoms. Consequently, papers in high-impact journals have called for identifying molecules and the development of drugs to directly inhibit CGRP 45 , which has since led to the development of CGRP inhibitors as a new class of migraine treatment medications.
Bipartite author-entity network
A total of 28,223 disambiguated authors and 5,379 distinct bio-entities of coronavirus articles were used to construct author-bio-entity bipartite network. Figure 5 illustrated the bipartite network (Fig. 5(a) ) and its author projection (Fig. 5(b) ) and bio-entity projection (Fig. 5(c) ). In Fig. 5(a) , the author vertices are blue, and the bio-entity vertices are pink. A link between a bio-entity and an author exists if and only if this bio-entity has been researched by that author. Connections between two authors or between two bio-entities are not allowed. The edge weight is set as the number of papers an author published that mention a bio-entity. In Fig. 5(b,c) , the edge weight is set as the number of common neighbors for the author and bio-entity, respectively. Vertices are marked with different colors to show their community attribution.
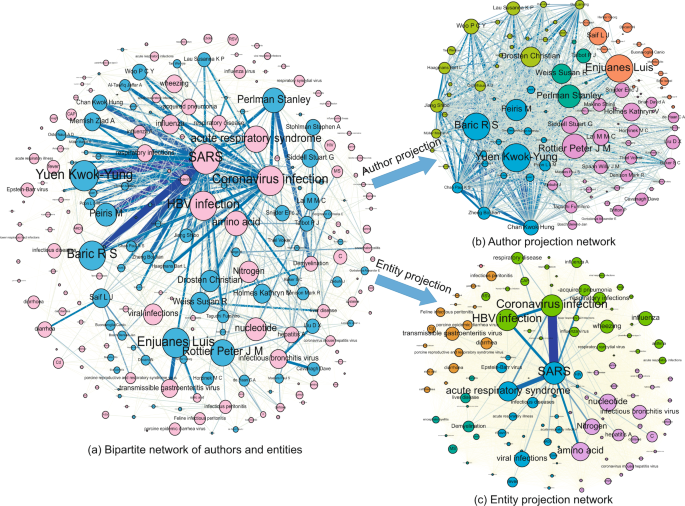
Bipartite network analysis of coronavirus.
Figure 5(a) illustrates a distinct relationship between authors and their focused bio-entities. For example, the disease SARS have been frequently studied by author Baric R S, Yuen Kwok-Yung, and Zheng Bo-Jian. In addition to SARS, Baric R S is also interested in coronavirus infection and HBV infection. Figure 5(b) depicts the common research interest relationship between authors. Strong connections between authors may indicate that they collaborated multiple times, such as Chan Kwok Hung and Yuen Kwok-Yung, who published 69 papers together. These connections may also indicate author pairs that have similar research interests but never collaborated, such as Baric R S and Yuen Kwok-Yung, which is crucial for the collaborative commendation. Similarly, the connections between bio-entities in Fig. 5(c) indicate that they have been studied by authors with similar research interests, which can be further applied to discover the hidden relations between bio-entities.
Code availability
We have made the pre-trained weights of BioBERT freely available at https://github.com/naver/biobert-pretrained , and the source code for fine-tuning BioBERT available at https://github.com/dmis-lab/biobert .
Hakala, K., Kaewphan, S., Salakoski, T. & Ginter, F. Syntactic analyses and named entity recognition for PubMed and PubMed Central—up-to-the-minute. In Proceedings of the 15th Workshop on Biomedical Natural Language Processing 102–107, https://doi.org/10.18653/v1/W16-2913 (2016).
Bell, L., Chowdhary, R., Liu, J. S., Niu, X. & Zhang, J. Integrated bio-entity network: a system for biological knowledge discovery. PLoS One 6 , e21474 (2011).
Article ADS CAS PubMed PubMed Central Google Scholar
Torvik, V. I. MapAffil: a bibliographic tool for mapping author affiliation strings to cities and their geocodes worldwide. Dlib Mag. 21 , 11–12, https://doi.org/10.1045/november2015-torvik (2015).
Article Google Scholar
Achakulvisut T. Affiliation parser. GitHub , https://github.com/titipata/affiliation_parser/wiki (2017).
Torvik, V. I. & Smalheiser, N. R. Author name disambiguation in MEDLINE. ACM Trans. Knowl. Discov. Data 3 , 11, https://doi.org/10.1145/1552303.1552304 (2009).
Article PubMed PubMed Central Google Scholar
Blackburn, R. et al . ORCID Public Data File 2018. figshare https://doi.org/10.23640/07243.7234028.v1 (2018).
Lee, J. et al . BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics 36 , 1234–1240, https://doi.org/10.1093/bioinformatics/btz682 (2019).
Article CAS PubMed Central Google Scholar
Kim, D. et al . A neural named entity recognition and multi-type normalization tool for biomedical text mining. IEEE Access 7 , 73729–73740 (2019).
Ammar, W. et al . Construction of the literature graph in semantic scholar. In Proceedings of the 2018 Conference of the NAACH-HLT 3 , 84–91, https://doi.org/10.18653/v1/N18-3011 (2018).
NIH. NIH ExPORTER dataset 2018, http://exporter.nih.gov (2018).
Torvik, V. I. MapAffil 2016 dataset–PubMed author affiliations mapped to cities and their geocodes worldwide. University of Illinois at Urbana-Champaign , https://doi.org/10.13012/B2IDB-4354331_V1 (2018).
Habibi, M. et al . Deep learning with word embeddings improves biomedical named entity recognition. Bioinformatics 33 , i37–i48 (2017).
Article CAS PubMed PubMed Central Google Scholar
Yoon, W., So, C. H., Lee, J. & Kang, J. CollaboNet: collaboration of deep neural networks for biomedical named entity recognition. BMC Bioinformatics 20 , 249 (2019).
Devlin, J., Chang, M. W., Lee, K. & Toutanova, K. Bert: pre-training of deep bidirectional transformers for language understanding. In Proceedings of the 2019 Conference of the NAACH-HLT 1 , 4171–4186, https://doi.org/10.18653/v1/N19-1423 (2019).
Wu, Y. et al . Google’s neural machine translation system: bridging the gap between human and machine translation. Preprint at, https://arxiv.org/abs/1609.08144 (2016).
Sang, E. F. & Veenstra, J. Representing text chunks. In Proceedings of the Ninth Conference on EACL 173–179, https://doi.org/10.3115/977035.977059 (1999).
Buchholz, S. & Marsi, E. CoNLL-X shared task on multilingual dependency parsing. In Proceedings of the Tenth Conference on CoNLL . ACL 149–164, https://doi.org/10.5555/1596276.1596305 (2006).
Law, V. et al . DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res. 42 , D1091–D1097 (2013).
Article PubMed PubMed Central CAS Google Scholar
Li, J. C., Yin, Y., Fortunato, S. & Wang, D. S. A dataset of publication records for Nobel laureates. Scientific Data 6 , 33 (2019).
Laudel, G. Studying the brain drain: can bibliometric methods help? Scientometrics 57 , 215–237 (2003).
Article CAS Google Scholar
Liu, W. et al . Author name disambiguation for PubMed. J. Assoc. Inf. Sci. Tech. 65 , 765–781 (2014).
Wu, J. & Ding, X. H. Author name disambiguation in scientific collaboration and mobility cases. Scientometrics 96 , 683–697 (2013).
Kang, I. S. et al . On co-authorship for author disambiguation. Inf. Process. Manage. 45 , 84–97 (2009).
Levin, M., Krawczyk, S., Bethard, S. & Jurafsky, D. Citation‐based bootstrapping for large‐scale author disambiguation. J. Am. Soc. Inf. Sci. Technol. 63 , 1030–1047 (2012).
Wu, H., Li, B., Pei, Y. J. & He, J. Unsupervised author disambiguation using Dempster–Shafer theory. Scientometrics 101 , 1955–1972 (2014).
Shin, D., Kim, T., Choi, J. & Kim, J. Author name disambiguation using a graph model with node splitting and merging based on bibliographic information. Scientometrics 100 , 15–50 (2014).
ORCID. About ORCID , https://orcid.org/about (2019).
NLM. MEDLINE PubMed XML element descriptions and their attributes, https://www.nlm.nih.gov/bsd/licensee/elements_descriptions.html#meshheadinglist (2019).
Xu, J. et al . Building a PubMed knowledge graph. figshare https://doi.org/10.6084/m9.figshare.c.4773944 (2020).
NLM. Download MEDLINE/PubMed Data, https://www.nlm.nih.gov/databases/download/pubmed_medline.html (2019).
Sachan, D. S., Xie, P. T., Sachan, M. & Xing, E. P. Effective use of bidirectional language modeling for transfer learning in biomedical named entity recognition. In Machine Learning for Healthcare Conference 85 , 1–19, http://proceedings.mlr.press/v85/sachan18a/sachan18a.pdf (2018).
Zhu, H., Paschalidis, I. C. & Tahmasebi, A. Clinical concept extraction with contextual word embedding. In NIPS Machine Learning for Health Workshop 1–6, https://arxiv.org/abs/1810.10566 (2018).
Wang, X. et al . Cross-type biomedical named entity recognition with deep multi-task learning. Bioinformatics 35 , 1745–1752 (2019).
Article CAS PubMed Google Scholar
Giorgi, J. M. & Bader, G. D. Transfer learning for biomedical named entity recognition with neural networks. Bioinformatics 34 , 4087–4094 (2018).
Lerchenmueller, M. J. & Sorenson, O. Author disambiguation in PubMed: evidence on the precision and recall of author-ity among NIH-funded scientists. PLoS One 11 , e0158731 (2016).
Kawashima, H. & Tomizawa, H. Accuracy evaluation of Scopus Author ID based on the largest funding database in Japan. Scientometrics 103 , 1061–1071 (2015).
Warner, E. T., Carapinha, R., Weber, G. M., Hill, E. V. & Reede, J. Y. Faculty promotion and attrition: the importance of coauthor network reach at an academic medical center. J. Gen. Intern. Med. 31 , 60–67 (2016).
Article PubMed Google Scholar
Griffin, M. Professional networking and expertise mining for research collaboration. Profiles research networking software , http://profiles.catalyst.harvard.edu/?pg=home (2019).
ELSEVIER. Elsevier fingerprint engine , https://www.elsevier.com/solutions/elsevier-fingerprint-engine (2019).
CUSP. CUSP scientific profiles , https://cusp.irvinginstitute.columbia.edu/cusp/cgi-bin/ww2ui.cgi/splash (2019).
UCI. Discover UCI faculty , https://www.faculty.uci.edu/ (2019).
Yue, W., Yang, C. S., DiPaola, R. S. & Tan, X. L. Repurposing of metformin and aspirin by targeting AMPK-mTOR and inflammation for pancreatic cancer prevention and treatment. Cancer Prev. Res. 7 , 388–397 (2014).
Bertolini, F., Sukhatme, V. P. & Bouche, G. Drug repurposing in oncology—patient and health systems opportunities. Nat. Rev. Clin. Oncol. 12 , 732–742 (2015).
Durham, P. L. Calcitonin gene‐related peptide (CGRP) and migraine. Headache 46 , S3–S8 (2006).
Durham, P. L. CGRP-receptor antagonists—a fresh approach to migraine therapy? N. Engl. J. Med. 350 , 1073–1075 (2004).
Maglott, D., Ostell, J., Pruitt, K. D. & Tatusova, T. Entrez gene: gene-centered information at NCBI. Nucleic Acids Res. 39 , D52–D57, https://doi.org/10.1093/nar/gkq1237 (2010).
D’Souza, J. & Ng, V. Sieve-based entity linking for the biomedical domain. In Proceedings of AACL-IJCNLP 2015 2 , 297–302, https://doi.org/10.3115/v1/P15-2049 (2015).
Lipscomb, C. E. Medical subject headings (MeSH). Bull. Med. Libr. Assoc. 88 , 265–266 (2000).
CAS PubMed PubMed Central Google Scholar
Hamosh, A., Scott, A. F., Amberger, J. S., Bocchini, C. A. & McKusick, V. A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 33 , D514–D517 (2005).
Donnelly, K. SNOMED-CT: the advanced terminology and coding system for eHealth. Stud. Health Tech. Informat. 121 , 279 (2006).
Google Scholar
Liu, Y. F., Liang, Y. J. & Wishart, D. PolySearch2: a significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. 43 , W535–W542 (2015).
Degtyarenko, K. et al . ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 36 , D344–D350 (2007).
Sherry, S. T. et al . dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29 , 308–311 (2001).
Landrum, M. J. et al . ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44 , D862–D868 (2016).
Doğan, R. I., Leaman, R. & Lu, Z. Y. NCBI disease corpus: a resource for disease name recognition and concept normalization. J. Biomed. Inform. 47 , 1–10 (2014).
Uzuner, Ö., South, B. R., Shen, S. Y. & DuVall, S. L. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J. Am. Med. Inf. Assoc. 18 , 552–556 (2011).
Li, J. et al . BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database(Oxford) 2016 , baw068, https://doi.org/10.1093/database/baw068 (2016).
Krallinger, M. et al . The CHEMDNER corpus of chemicals and drugs and its annotation principles. J. Cheminformatics 7 , S2 (2015).
Smith, L. et al . Overview of BioCreative II gene mention recognition. Genome Biol. 9 , S2 (2008).
Kim, J. D., Ohta, T., Tsuruoka, Y., Tateisi, Y. & Collier, N. Introduction to the bio-entity recognition task at JNLPBA. In Proceedings of the NLPBA/BioNLP . ACL 70–75, https://doi.org/10.3115/1567594.1567610 (2004).
Gerner, M., Nenadic, G. & Bergman, C. M. LINNAEUS: a species name identification system for biomedical literature. BMC Bioinformatics 11 , 85 (2010).
Pafilis, E. et al . The SPECIES and ORGANISMS resources for fast and accurate identification of taxonomic names in text. PLoS One 8 , e65390 (2013).
Morgan, A. A. et al . Overview of BioCreative II gene normalization. Genome Biol. 9 , S3 (2008).
Lu, Z. et al . The gene normalization task in BioCreative III. BMC Bioinformatics 12 , S2 (2011).
Pradhan, S. et al . Task 1: ShARe/CLEF eHealth Evaluation Lab. CLEF 1–6, https://pdfs.semanticscholar.org/7dfb/97a2b878673e67062eeab0ba1871eae9a893.pdf (2013).
Furlong, L. I., Dach, H., Hofmann-Apitius, M. & Sanz, F. OSIRISv1. 2: a named entity recognition system for sequence variants of genes in biomedical literature. BMC Bioinformatics 9 , 84 (2008).
Thomas, P. E., Klinger, R., Furlong, L. I., Hofmann-Apitius, M. & Friedrich, C. M. Challenges in the association of human single nucleotide polymorphism mentions with unique database identifiers. BMC Bioinformatics 12 , S4 (2011).
Wei, C. H., Kao, H. Y. & Lu, Z. SR4GN: a species recognition software tool for gene normalization. PLoS One 7 , e38460 (2012).
Carroll, H. D. et al . Threshold Average Precision (TAP-k): a measure of retrieval designed for bioinformatics. Bioinformatics 26 , 1708–1713 (2010).
Download references
Acknowledgements
This work was supported by National Social Science Fund of China [18BTQ076], Chinese National Youth Foundation Research [61702564], Natural Science Foundation of Guangdong Province [2018A030313981], Soft Science Foundation of Guangdong Province [2019A101002020], National Research Foundation of Korea [NRF-2019R1A2C2002577] and [NRF-2017R1A2A1A17069645], and US National Institutes of Health [P01AG039347]. The authors acknowledge the Texas Advanced Computing Center (TACC) at The University of Texas at Austin for providing storage resources that have contributed to the research results reported within this paper. URL: http://www.tacc.utexas.edu .
Author information
Authors and affiliations.
School of Information Management, Sun Yat-sen University, Guangzhou, China
Jian Xu & Daifeng Li
Department of Computer Science and Engineering, Korea University, Seoul, South Korea
Sunkyu Kim, Minbyul Jeong, Donghyeon Kim & Jaewoo Kang
Department of Library and Information Science, Yonsei University, Seoul, South Korea
Dell Medical School, University of Texas at Austin, Austin, TX, USA
Justin F. Rousseau & Ying Ding
School of Information, University of Texas at Austin, Austin, TX, USA
Xin Li, Chongyan Chen, Islam Akef Ebeid & Ying Ding
Texas Advanced Computing Center, Austin, TX, USA
School of Information Sciences, University of Illinois at Urbana-Champaign, Champaign, IL, USA
Vetle I. Torvik
Department of Information Management, Peking University, Beijing, China
You can also search for this author in PubMed Google Scholar
Contributions
Y.D., J.X. and D.L. proposed the idea and supervised the project. J.X., Y.D. and M.S. wrote and revised this manuscript. S.K., M.J., D.K. and J.K. conducted the bio-entity extraction and validity. J.R., X.L., W.X., Y.B., C.C. and I.A.E. conducted the usage notes. V.I.T. and M.S. conducted the author name disambiguation and validity.
Corresponding authors
Correspondence to Daifeng Li or Ying Ding .
Ethics declarations
Competing interests.
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ .
The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files associated with this article.
Reprints and permissions
About this article
Cite this article.
Xu, J., Kim, S., Song, M. et al. Building a PubMed knowledge graph. Sci Data 7 , 205 (2020). https://doi.org/10.1038/s41597-020-0543-2
Download citation
Received : 11 December 2019
Accepted : 26 May 2020
Published : 26 June 2020
DOI : https://doi.org/10.1038/s41597-020-0543-2
Share this article
Anyone you share the following link with will be able to read this content:
Sorry, a shareable link is not currently available for this article.
Provided by the Springer Nature SharedIt content-sharing initiative
This article is cited by
What do we know about the disruption index in scientometrics an overview of the literature.
- Christian Leibel
- Lutz Bornmann
Scientometrics (2024)
Constructing a knowledge graph for open government data: the case of Nova Scotia disease datasets
- Enayat Rajabi
- Rishi Midha
- Jairo Francisco de Souza
Journal of Biomedical Semantics (2023)
Analysis and implementation of the DynDiff tool when comparing versions of ontology
- Sara Diaz Benavides
- Silvio D. Cardoso
- Cédric Pruski
Building a knowledge graph to enable precision medicine
- Payal Chandak
- Kexin Huang
- Marinka Zitnik
Scientific Data (2023)
From language models to large-scale food and biomedical knowledge graphs
- Gjorgjina Cenikj
- Lidija Strojnik
- Tome Eftimov
Scientific Reports (2023)
Quick links
- Explore articles by subject
- Guide to authors
- Editorial policies
Sign up for the Nature Briefing newsletter — what matters in science, free to your inbox daily.
- Databases for Keyword Searches
- Login to databases
- Guides by Librarians
- Advanced Search
- Renew a book
- Journals by Name
- Textbooks & Course Reserves

- Textbooks & Course Reserves
Statistics, Data, Graphs, and Other Factual Support: Citing a Graph with Data
- Using These Statistics
- General Statistics
- Children & Education
- Demographics
- Environment
- Health & Safety
- Human Rights
- International Statistics
- Politics & Economy
- Sports & Recreation
- Transportation
- Citing a Graph with Data
Citing a Photo, Image, Graph, or Chart with MLA
Cite a graph.
The rules for citing a graph are the same for citing a photo, illustration, map, or diagram. Place the image in the body of the essay where it is pertinent to the subject matter, and give the citation after labeling it with "Fig." and a number. Use the numbers consecutively from 1 on.

Fig. 1. Party agree graph from: McCready, Ryan. "5 Ways Writers Use Misleading Graphs To Manipulate You [INFOGRAPHIC]." Venngage, 9 Sep 2018.
Continue to Use Double Spacing
The citation should be double spaced and the only difference should be the notation "Fig." and the number which should be bold with periods after the notation and number as shown here.

Using Statistics in a Paper
- Writing and Reading Statistics A guide from University of North Carolina's Writing Center on using statistics to make your argument useful.
- Writing with statistics From the OWL Purdue Website
- << Previous: Transportation
- Last Updated: Jul 20, 2023 8:57 AM
- URL: https://shoreline.libguides.com/statistics
University Libraries University of Nevada, Reno
- Skill Guides
- Subject Guides
MLA Citation Guide (MLA 9th Edition): Charts, Graphs, Images, and Tables
- Understanding Core Elements
- Formatting Appendices and Works Cited List
- Writing an Annotated Bibliography
- Academic Honesty and Citation
- In-Text Citation
- Charts, Graphs, Images, and Tables
- Class Notes and Presentations
- Encyclopedias and Dictionaries
- Generative AI
- In Digital Assignments
- Interviews and Emails
- Journal and Magazine Articles
- Newspaper Articles
- Social Media
- Special Collections
- Videos and DVDs
- When Information Is Missing
- Citation Software
Is it a Figure or a Table?
There are two types of material you can insert into your assignment: figures and tables. A figure is a photo, image, map, graph, or chart. A table is a table of information. For a visual example of each, see the figure and table to the right.
Still need help? For more information on citing figures, visit Purdue OWL .
Reproducing Figures and Tables
Reproducing happens when you copy or recreate a photo, image, chart, graph, or table that is not your original creation. If you reproduce one of these works in your assignment, you must create a note (or "caption") underneath the photo, image, chart, graph, or table to show where you found it. If you do not refer to it anywhere else in your assignment, you do not have to include the citation for this source in a Works Cited list.
Citing Information From a Photo, Image, Chart, Graph, or Table
If you refer to information from the photo, image, chart, graph, or table but do not reproduce it in your paper, create a citation both in-text and on your Works Cited list.
If the information is part of another format, for example a book, magazine article, encyclopedia, etc., cite the work it came from. For example if information came from a table in an article in National Geographic magazine, you would cite the entire magazine article.
Figure Numbers
The word figure should be abbreviated to Fig. Each figure should be assigned a figure number, starting with number 1 for the first figure used in the assignment. E.g., Fig. 1.
Images may not have a set title. If this is the case give a description of the image where you would normally put the title.
A figure refers to a chart, graph, image or photo. This is how to cite figures.
The caption for a figure begins with a description of the figure followed by the complete citation for the source the figure was found in. For example, if it was found on a website, cite the website. If it was in a magazine article, cite the magazine article.
- Label your figures starting at 1.
- Information about the figure (the caption) is placed directly below the image in your assignment.
- If the image appears in your paper the full citation appears underneath the image (as shown below) and does not need to be included in the Works Cited List. If you are referring to an image but not including it in your paper you must provide an in-text citation and include an entry in the Works Cited.

Fig. 1. Man exercising from: Green, Annie. "Yoga: Stretching Out." Sports Digest, 8 May 2006, p. 22.

Fig. 2. Annakiki skirt from: Cheung, Pauline. "Short Skirt S/S/ 15 China Womenswear Commercial Update." WGSN.
Images: More Examples
In the works cited examples below, the first one is seeing the artwork in person, the second is accessing the image from a website, the third is accessing it through a database, and the last example is using an image from a book.
Viewing Image in Person
Hopper, Edward. Nighthawks . 1942, Art Institute of Chicago.
Accessing Image from a Website
Hopper, Edward. Nighthawks . 1942. Art Institute of Chicago, www.artic.edu/aic/collections/artwork/111628 .
Note : Notice the period after the date in the example above, rather than a comma as the other examples use. This is because the date refers to the painting's original creation, rather than to its publication on the website. It is considered an "optional element."
Accessing Image from a Database
Hopper, Edward. Nighthawks . 1942, Art Institute of Chicago. Artstor , https://library.artstor.org/#/asset/AWSS35953_35953_41726475 .
Using an Image from a Book
Hopper, Edward. Nighthawks . 1942, Art Institute of Chicago. Staying Up Much Too Late: Edward Hopper's Nighthawks and the Dark Side of the American Psyche , by Gordon Theisen, Thomas Dunne Books, 2006, p. 118.
Above the table, label it beginning at Table 1, and add a description of what information is contained in the table.
The caption for a table begins with the word Source, then the complete Works Cited list citation for the source the table was found in. For example, if it was found on a website, cite the website. If it was in a journal article, cite the journal article.
Information about the table (the caption) is placed directly below the table in your assignment.
If the table is not cited in the text of your assignment, you do not need to include it in the Works Cited list.
Variables in determining victims and aggressors
Source: Mohr, Andrea. "Family Variables Associated With Peer Victimization." Swiss Journal of Psychology, vol . 65, no. 2, 2006, pp. 107-116. Psychology Collection , doi: http://dx.doi.org/10.1024/1421-0185.65.2.107.
- << Previous: Books
- Next: Class Notes and Presentations >>

APA 7th Referencing
- Style summary
- Easy Referencing tool This link opens in a new window
- In-text citations
- Reference lists
- Secondary sources (as cited in)
- Streaming videos
- Film/Movie, TV, radio and podcasts
- Print books
- Book chapters
- Edited books
- Conference papers and webinars
- Dictionaries and encyclopedias
- First Nations resources and knowledges
- Images, artworks, and screenshots
- Journal articles
- Newspapers and magazines
- Lecture/Class materials, MOOCs/learning modules and personal communications
- Legal cases
- Legislation, bills and regulations
- Conventions and treaties
- Taxation rulings
- Medical databases
- Plant labels and profiles
- Standards, building codes and patents
- Graphs (figures)
Graphs (figures) format
- Theses and dissertations
- Translated and foreign works
- Websites and webpages
- Online documents (e.g. white paper, brochure, fact sheet, ppt slides etc.)
- Social media, apps, games and AI
- APA 7th quiz
- From website
- From database
- From electronic journal
Referencing your own graphs/figures
Graph/figure from a website , in-text citation.
More examples of website references.
Graph/Figure from a database
Graph/figure from a book, graph/figure from an electronic journal.
More example of ejournal references .
If the table is unpublished, you do not need to include a reference (see: personal communications tab ).
- << Previous: Tables
- Next: Theses and dissertations >>
- Last Updated: Apr 2, 2024 3:09 PM
- URL: https://holmesglen.libguides.com/apa7
Citation guides
All you need to know about citations
How to cite a graph in MLA
It is common practice to cite the work the graph has been published in and provide the page number in the in-text citation. In case the graph has not been published in a journal article, book, or book chapter, but is rather found online take a look at our MLA photo citation guides below.
MLA citation format for a graph
- Google Docs
To cite a graph in a reference entry in MLA style 8th edition include the following elements:
- Author: Give the last name and name as presented in the source (e. g. Watson, John). For two authors, reverse only the first name, followed by ‘and’ and the second name in normal order (e. g. Watson, John, and John Watson). For three or more authors, list the first name followed by et al. (e. g. Watson, John, et al.)
- Title of the graph: Titles are italicized when independent. If part of a larger source add quotation marks and do not italize.
- Year of publication: Give the year of publication as presented in the source.
- Title of website: If the name of an academic press contains the words University and Press, use UP e.g. Oxford UP instead of Oxford University Press. If the word "University" doesn't appear, spell out the Press e.g. MIT Press.
- URL: Copy URL in full from your browser, include http:// or https:// and do not list URLs created by shortening services.
Here is the basic format for a reference list entry of a graph in MLA style 8th edition:
Author . Title of the graph . Year of publication . Title of website , URL .
- Author(s) of the book: Give the last name and name as presented in the source (e. g. Watson, John). For two authors, reverse only the first name, followed by ‘and’ and the second name in normal order (e. g. Watson, John, and John Watson). For three or more authors, list the first name followed by et al. (e. g. Watson, John, et al.)
- Title of the book:
- Publisher: If the name of an academic press contains the words University and Press, use UP e.g. Oxford UP instead of Oxford University Press. If the word "University" doesn't appear, spell out the Press e.g. MIT Press.
Author(s) of the book . Title of the book Publisher , Year of publication .
Take a look at our works cited examples that demonstrate the MLA style guidelines in action:
Graph citation from a digital source
Masoud, Carla . Social media usage in young adults . 2017 . Psych Publish , psychology-now.org/graphs/social-media-stats/ .
Graph citation from a book
Devito, Roberto . Cheese consumption in the USA . Chicago Publishing , 2021 .
How to do an in-text citation for a graph in MLA
When citing a graph in-text using the MLA style, you'll use the surname of the creator followed by the page number in parentheses.
In practice, you can expect your graph's in-text citation to be in this format (Author, Page Number) .
If you were to cite a graph from a book, the graph should be cited in-text using the creator's name, along with the corresponding year of publication.
Citation of a graph from a book on page 193
Survey showed that 80% of high-school students were sleep-deprived (Eid, 193) .
If the creator is not mentioned, you can place the graph's title or description instead.
Citation of a graph from a source with no creator
Zinc was found to be one of the most prevalent heavy metals in the Nile River ("Levels of heavy metals in the Nile River", 198) .
If the graph is found online, do not list a page number.
Citation of a graph found online
It is estimated that 60% of start-ups go bankrupt in the first 10 years (Eid) .

This citation style guide is based on the MLA Handbook (9 th edition).
More useful guides
- Citing Images in MLA 8th Edition
- MLA Style Center Citing online images
More great BibGuru guides
- MLA: how to cite a book chapter
- AMA: how to cite a software manual
- MLA: how to cite a PhD thesis
Automatic citations in seconds
Citation generators
Alternative to.
- NoodleTools
- Getting started
From our blog
- 📚 How to write a book report
- 📝 APA Running Head
- 📑 How to study for a test

IMAGES
VIDEO
COMMENTS
Get a visual overview of a new academic field. Enter a typical paper and we'll build you a graph of similar papers in the field. Explore and build more graphs for interesting papers that you find - soon you'll have a real, visual understanding of the trends, popular works and dynamics of the field you're interested in.
Note: This page reflects the latest version of the APA Publication Manual (i.e., APA 7), which released in October 2019. The equivalent resources for the older APA 6 style can be found at this page as well as at this page (our old resources covered the material on this page on two separate pages). The purpose of tables and figures in documents is to enhance your readers' understanding of the ...
To cite a graph in MLA style, refer to the graph in the text as Figure 1 in parentheses, and place a caption under the graph that says "Figure 1." Then, include a short description, such as the title of the graph, and list the authors first and last name, as well as the publication name, with the location, publisher, and year in parentheses.
All three groups of identified papers can then be exported into most reference managers like Zotero, EndNote, Mendeley. For any paper identified of interest you can further click on the "build a graph" button to make it the seed paper instead and generate yet another similarity graph using it as a seed paper.
The citation graph, or citation network, is a model used to describe how citations link research entities, typically papers, journals, and books (Harzing & Van der Wal, 2008; Tang et al., 2008).It enables a number of important activities such as the following: Exploration of the graph to find publications of interest.. Tracking of authorship of papers: Citing and following citations is one way ...
By activating the Connected Papers toggle switch, readers can follow a link to the article's graph displayed at Connected Papers. Each paper's graph is created by analyzing tens of thousands of papers for similarity in their citations, and then a small subset of those analyzed are arranged according to their degree of similarity. ...
Connected Papers is a tool for thought to help researchers and applied scientists find and explore papers relevant to their field of work in a visual way. You enter an origin paper, and they generate a graph. To achieve this, they analyse about 50,000 research papers, and select the ones with the strongest connections to the origin paper.
How to use. Citationtree searches for papers in the citation environment of the input then displays only the most central papers. The darker nodes are previous papers at a distance 1 from the input, the lighter nodes are previous papers at a distance 2 from the input and papers that cite the input. Export the tree as SVG, RIS or BibTeX using ...
Citegraph contains 32+ million paper citation edges. Fun fact: Distinctive Image Features from Scale-Invariant Keypoints is the most cited paper. Author ---WRITES--> Paper. Citegraph contains 16+ million authorship edges. Fun fact: H. V. POOR is the most productive researcher - he has authored more than 1.6k papers!
Tables and figures taken from other sources are numbered and presented in the same format as your other tables and figures. Refer to them as Table 1, Figure 3, etc., but include an in-text citation after you mention them to acknowledge the source. In-text citation example. The results in Table 1 (Ajzen, 1991, p. 179) show that ….
Citing Information From an Image, Chart, Table or Graph. If you refer to information from an image, chart, table or graph, but do not reproduce it in your paper, create a citation both in-text and on your Reference list. If the information is part of another format, for example a book, magazine article, encyclopedia, etc., cite the work it came ...
A figure may be a chart, a graph, a photograph, a drawing, or any other illustration or nontextual depiction. Any type of illustration or image other than a table is referred to as a figure. Figure Components. Number: The figure number (e.g., Figure 1) appears above the figure in bold (no period finishing). Title: The figure title appears one double-spaced line below the figure number in ...
This happens if you only cite information from an image, infographic, chart, table, or graph and do not reproduce it in your paper. If you're only citing information from an image, infographic, Chart, Table or Graph: Provide an in-text citation. Use the citation format of the source where the image is found.
In-Text Citation (Quotation) - entry that appears in the body of your paper after a direct quote. References - entry that appears at the end of your paper. Figures. When you use a figure in your paper that has been adapted or copied directly from another source, you need to reference the original source. This reference appears as a caption ...
Placement of figures in a paper. There are two options for the placement of figures (and tables) in a paper. The first is to embed figures in the text after each is first mentioned (or "called out"); the second is to place each figure on a separate page after the reference list. An embedded figure may take up an entire page; if the figure ...
Scientific Reports (2023) PubMed® is an essential resource for the medical domain, but useful concepts are either difficult to extract or are ambiguous, which has significantly hindered knowledge ...
Cite a Graph. The rules for citing a graph are the same for citing a photo, illustration, map, or diagram. Place the image in the body of the essay where it is pertinent to the subject matter, and give the citation after labeling it with "Fig." and a number. Use the numbers consecutively from 1 on.
A figure refers to a chart, graph, image or photo. This is how to cite figures. The caption for a figure begins with a description of the figure followed by the complete citation for the source the figure was found in. For example, if it was found on a website, cite the website. If it was in a magazine article, cite the magazine article.
Figure #. Figure title (Author, Year) [Graph/figure goes below the heading] Figure 1. Number of people who took part in the magic penguin dance in 2018 (Bookman, 2019) Format. Example. Refer to figures according to how you've numbered them (i.e. Figure 1, Figure 2, etc.) As shown in Figure 1 ...
Figures include diagrams and all types of graphs. An i m a ge, photo, illustration or screenshot displayed for scientific purposes is classed as a figure.. All figures in your paper must be referred to in the main body of the text. At the bottom of the figure is the title, explaining what the figure is showing and the legend, i.e. an explanation of what the symbols, acronyms or colours mean.
Don't include an entry in the reference list. Personal communication may include materials such as emails from unarchived sources, private memos or unrecorded interview conversations. Confidential material may include medical charts, patient health records and other internal reports containing private information. ...
Online. In a book. To cite a graph in a reference entry in MLA style 8th edition include the following elements: Author: Give the last name and name as presented in the source (e. g. Watson, John). For two authors, reverse only the first name, followed by 'and' and the second name in normal order (e. g. Watson, John, and John Watson).
Scene Graph Generation (SGG) is a challenging task of detecting objects and predicting relationships between objects. After DETR was developed, one-stage SGG models based on a one-stage object detector have been actively studied. However, complex modeling is used to predict the relationship between objects, and the inherent relationship between object queries learned in the multi-head self ...
Dating app Tinder announced a new April Fools' Day hiring quest for a Vice President of Ghost Hunting to help combat "one of dating culture's most prevalent vices - ghosting," a practice ...